[English] 日本語
 Yorodumi
Yorodumi- EMDB-9040: Analysis of bacteriophage phiX174 tomograms using the locked self... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9040 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Analysis of bacteriophage phiX174 tomograms using the locked self-rotation function | ||||||||||||
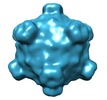
 Map data Map data | The map was calculated by averaging of ten icosahedrally-averaged maps of individual phiX174 particles. | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Enterobacteria phage phiX174 (virus) Enterobacteria phage phiX174 (virus) | ||||||||||||
| Method | electron tomography / cryo EM | ||||||||||||
 Authors Authors | Fokine A / Sun Y / Khare B / Rossmann MG | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2019 Journal: J Struct Biol / Year: 2019Title: Enhancement of tomogram interpretability using the locked self-rotation function. Authors: Andrei Fokine / Baldeep Khare / Yingyuan Sun / Michael G Rossmann /  Abstract: The interpretation of cryo-electron tomograms of macromolecular complexes can be difficult because of the large amount of noise and because of the missing wedge effect. Here it is shown how the ...The interpretation of cryo-electron tomograms of macromolecular complexes can be difficult because of the large amount of noise and because of the missing wedge effect. Here it is shown how the presence of rotational symmetry in a sample can be utilized to enhance the quality of a tomographic analysis. The orientation of symmetry axes in a sub-tomogram can be determined using a locked self-rotation function. Given this knowledge, the sub-tomogram density can then be averaged to improve its interpretability. Sub-tomograms of the icosahedral bacteriophage phiX174 are used to demonstrate the procedure. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9040.map.gz emd_9040.map.gz | 474.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9040-v30.xml emd-9040-v30.xml emd-9040.xml emd-9040.xml | 33.7 KB 33.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9040.png emd_9040.png | 106.8 KB | ||
| Others |  emd_9040_additional_1.map.gz emd_9040_additional_1.map.gz emd_9040_additional_10.map.gz emd_9040_additional_10.map.gz emd_9040_additional_11.map.gz emd_9040_additional_11.map.gz emd_9040_additional_2.map.gz emd_9040_additional_2.map.gz emd_9040_additional_3.map.gz emd_9040_additional_3.map.gz emd_9040_additional_4.map.gz emd_9040_additional_4.map.gz emd_9040_additional_5.map.gz emd_9040_additional_5.map.gz emd_9040_additional_6.map.gz emd_9040_additional_6.map.gz emd_9040_additional_7.map.gz emd_9040_additional_7.map.gz emd_9040_additional_8.map.gz emd_9040_additional_8.map.gz emd_9040_additional_9.map.gz emd_9040_additional_9.map.gz | 1.8 MB 1.8 MB 518.5 KB 1.8 MB 1.8 MB 1.8 MB 1.8 MB 7.4 MB 1.8 MB 1.8 MB 1.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9040 http://ftp.pdbj.org/pub/emdb/structures/EMD-9040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9040 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9040.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9040.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map was calculated by averaging of ten icosahedrally-averaged maps of individual phiX174 particles. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
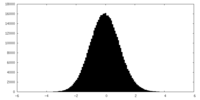
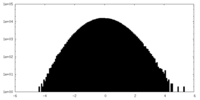
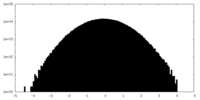
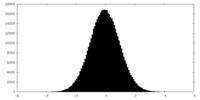
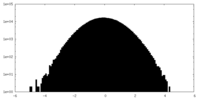
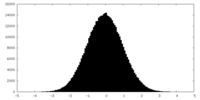
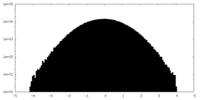
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
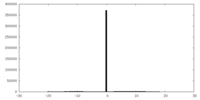
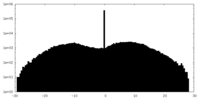
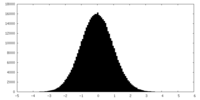
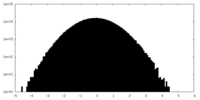
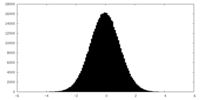
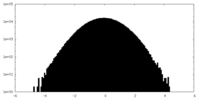
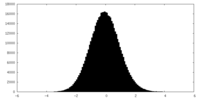
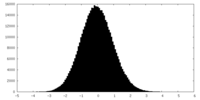
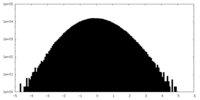
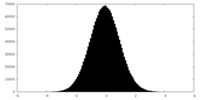
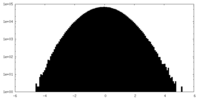
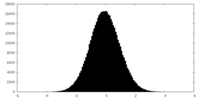
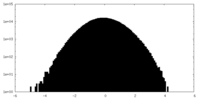
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: The map was produced by averaging of eight...
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: Tomogram of one bacteriophage phiX174 particle.
+Additional map: Tomogram of one bacteriophage phiX174 particle.
- Sample components
Sample components
-Entire : Enterobacteria phage phiX174
| Entire | Name:  Enterobacteria phage phiX174 (virus) Enterobacteria phage phiX174 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Enterobacteria phage phiX174
| Supramolecule | Name: Enterobacteria phage phiX174 / type: virus / ID: 1 / Parent: 0 / Details: The virus was grown in E.coli cells. / NCBI-ID: 10847 / Sci species name: Enterobacteria phage phiX174 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 320.0 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-2/2 4C / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 | ||||||||||||||
| Details | The concentration of the virus was 10**12 pfu/ml | ||||||||||||||
| Sectioning | Other: NO SECTIONING | ||||||||||||||
| Fiducial marker | Manufacturer: Aurion, Wageningen, Netherland / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.73 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: 3.0 µm / Nominal magnification: 11000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 41 IMOD / Number images used: 41 |
|---|
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)