+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8608 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of E. coli MCE protein PqiB, periplasmic domain | ||||||||||||
 Map data Map data | E. coli MCE protein PqiB, periplasmic domain | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | MCE protein / bacterial lipid transport / TRANSPORT PROTEIN | ||||||||||||
| Function / homology | : / Mce/MlaD / MlaD protein / intermembrane lipid transfer / membrane organization / outer membrane-bounded periplasmic space / identical protein binding / plasma membrane / Intermembrane transport protein PqiB Function and homology information Function and homology information | ||||||||||||
| Biological species |  | ||||||||||||
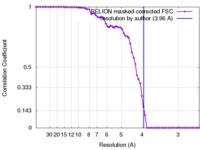
| Method | single particle reconstruction / cryo EM / Resolution: 3.96 Å | ||||||||||||
 Authors Authors | Bhabha G / Ekiert DC | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Architectures of Lipid Transport Systems for the Bacterial Outer Membrane. Authors: Damian C Ekiert / Gira Bhabha / Georgia L Isom / Garrett Greenan / Sergey Ovchinnikov / Ian R Henderson / Jeffery S Cox / Ronald D Vale /   Abstract: How phospholipids are trafficked between the bacterial inner and outer membranes through the hydrophilic space of the periplasm is not known. We report that members of the mammalian cell entry (MCE) ...How phospholipids are trafficked between the bacterial inner and outer membranes through the hydrophilic space of the periplasm is not known. We report that members of the mammalian cell entry (MCE) protein family form hexameric assemblies with a central channel capable of mediating lipid transport. The E. coli MCE protein, MlaD, forms a ring associated with an ABC transporter complex in the inner membrane. A soluble lipid-binding protein, MlaC, ferries lipids between MlaD and an outer membrane protein complex. In contrast, EM structures of two other E. coli MCE proteins show that YebT forms an elongated tube consisting of seven stacked MCE rings, and PqiB adopts a syringe-like architecture. Both YebT and PqiB create channels of sufficient length to span the periplasmic space. This work reveals diverse architectures of highly conserved protein-based channels implicated in the transport of lipids between the membranes of bacteria and some eukaryotic organelles. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8608.map.gz emd_8608.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8608-v30.xml emd-8608-v30.xml emd-8608.xml emd-8608.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8608_fsc.xml emd_8608_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_8608.png emd_8608.png | 78.5 KB | ||
| Filedesc metadata |  emd-8608.cif.gz emd-8608.cif.gz | 5.6 KB | ||
| Others |  emd_8608_additional.map.gz emd_8608_additional.map.gz | 3.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8608 http://ftp.pdbj.org/pub/emdb/structures/EMD-8608 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8608 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8608 | HTTPS FTP |
-Related structure data
| Related structure data |  5uvnMC  8610C  8611C 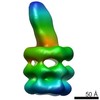 8612C  5uw2C  5uw8C  5uwaC  5uwbC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8608.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8608.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E. coli MCE protein PqiB, periplasmic domain | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Additional map, E. coli MCE protein PqiB, periplasmic domain
| File | emd_8608_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map, E. coli MCE protein PqiB, periplasmic domain | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : homo hexamer of PqiB
| Entire | Name: homo hexamer of PqiB |
|---|---|
| Components |
|
-Supramolecule #1: homo hexamer of PqiB
| Supramolecule | Name: homo hexamer of PqiB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 347 KDa |
-Macromolecule #1: Paraquat-inducible protein B
| Macromolecule | Name: Paraquat-inducible protein B / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.857043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHENL YFQSHQGPEV TLITANAEGI EGGKTTIKSR SVDVGVVESA TLADDLTHVE IKARLNSGME KLLHKDTVFW VVKPQIGRE GISGLGTLLS GVYIELQPGA KGSKMDKYDL LDSPPLAPPD AKGIRVILDS KKAGQLSPGD PVLFRGYRVG S VETSTFDT ...String: MHHHHHHENL YFQSHQGPEV TLITANAEGI EGGKTTIKSR SVDVGVVESA TLADDLTHVE IKARLNSGME KLLHKDTVFW VVKPQIGRE GISGLGTLLS GVYIELQPGA KGSKMDKYDL LDSPPLAPPD AKGIRVILDS KKAGQLSPGD PVLFRGYRVG S VETSTFDT QKRNISYQLF INAPYDRLVT NNVRFWKDSG IAVDLTSAGM RVEMGSLTTL LSGGVSFDVP EGLDLGQPVA PK TAFVLYD DQKSIQDSLY TDHIDYLMFF KDSVRGLQPG APVEFRGIRL GTVSKVPFFA PNMRQTFNDD YRIPVLIRIE PER LKMQLG ENADVVEHLG ELLKRGLRGS LKTGNLVTGA LYVDLDFYPN TPAITGIREF NGYQIIPTVS GGLAQIQQRL MEAL DKINK L(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) UniProtKB: Intermembrane transport protein PqiB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 20 mM Tris pH 8.0 and 150 mM NaCl |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 80.0 e/Å2 / Details: 80 e/A2 is total dose for 50 frames |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-5uvn: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)