+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8144 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Faustovirus | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus / double jelly-roll | |||||||||
| Function / homology | Group II dsDNA virus coat/capsid protein / Putative major capsid protein p72 Function and homology information Function and homology information | |||||||||
| Biological species |  Faustovirus Faustovirus | |||||||||
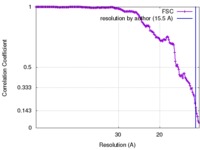
| Method | single particle reconstruction / cryo EM / Resolution: 15.5 Å | |||||||||
 Authors Authors | Klose T / Rossmann MG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2016 Journal: Proc Natl Acad Sci U S A / Year: 2016Title: Structure of faustovirus, a large dsDNA virus. Authors: Thomas Klose / Dorine G Reteno / Samia Benamar / Adam Hollerbach / Philippe Colson / Bernard La Scola / Michael G Rossmann /   Abstract: Many viruses protect their genome with a combination of a protein shell with or without a membrane layer. Here we describe the structure of faustovirus, the first DNA virus (to our knowledge) that ...Many viruses protect their genome with a combination of a protein shell with or without a membrane layer. Here we describe the structure of faustovirus, the first DNA virus (to our knowledge) that has been found to use two protein shells to encapsidate and protect its genome. The crystal structure of the major capsid protein, in combination with cryo-electron microscopy structures of two different maturation stages of the virus, shows that the outer virus shell is composed of a double jelly-roll protein that can be found in many double-stranded DNA viruses. The structure of the repeating hexameric unit of the inner shell is different from all other known capsid proteins. In addition to the unique architecture, the region of the genome that encodes the major capsid protein stretches over 17,000 bp and contains a large number of introns and exons. This complexity might help the virus to rapidly adapt to new environments or hosts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8144.map.gz emd_8144.map.gz | 113.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8144-v30.xml emd-8144-v30.xml emd-8144.xml emd-8144.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8144_fsc.xml emd_8144_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_8144.png emd_8144.png | 229.5 KB | ||
| Filedesc metadata |  emd-8144.cif.gz emd-8144.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8144 http://ftp.pdbj.org/pub/emdb/structures/EMD-8144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8144 | HTTPS FTP |
-Related structure data
| Related structure data |  5j7vMC  8145C  5j7oC  5j7uC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8144.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8144.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.58 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Faustovirus
| Entire | Name:  Faustovirus Faustovirus |
|---|---|
| Components |
|
-Supramolecule #1: Faustovirus
| Supramolecule | Name: Faustovirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1477405 / Sci species name: Faustovirus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Vermamoeba vermiformis (eukaryote) / Strain: CDC 19 Vermamoeba vermiformis (eukaryote) / Strain: CDC 19 |
| Virus shell | Shell ID: 1 / Name: outer capsid shell / Diameter: 2600.0 Å / T number (triangulation number): 277 |
| Virus shell | Shell ID: 2 / Name: inner capsid shell / Diameter: 1900.0 Å / T number (triangulation number): 16 |
-Macromolecule #1: major capsid protein
| Macromolecule | Name: major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Faustovirus Faustovirus |
| Molecular weight | Theoretical: 71.781891 KDa |
| Sequence | String: EAGGVFKLIA NDGKADRMIM ANDLLNDRIK SIMCLRAKQG FSDPTPTLVD IERTHILLIN SHYKPFAAMG YEYQKTRPNT GNPTYNSTI QFSIPQFGDF FSDMVVHVQL AATSASAGTV PALPAFIGAD DQVLTSTSVV SATENTTSGV YTLYTQSYVN Q QGTTQTVA ...String: EAGGVFKLIA NDGKADRMIM ANDLLNDRIK SIMCLRAKQG FSDPTPTLVD IERTHILLIN SHYKPFAAMG YEYQKTRPNT GNPTYNSTI QFSIPQFGDF FSDMVVHVQL AATSASAGTV PALPAFIGAD DQVLTSTSVV SATENTTSGV YTLYTQSYVN Q QGTTQTVA AAATNFVRYC EYPGLRLFKR VKFEVNGNPL DEYTALAAIM YNKFHVPDFK LTGWKRLIGQ EVPVEAASNL VN IASTTPW GSPIVALSDV NGTAVTGSPV NAAITARKLT QVVFGAQTPK ATQEQLNMFV PLLFWFRDPR LAIASVSIPY GQR FITVDI EQQSNILFTA PGNLFLQTTV ETLLTTGAGK GTATGVLLTQ YNRYTTYTPT LASGSSIDGT QAVQNIELYI NNIF VTPEI HDIYIKRIGF TLIRVYREQV QREVNAADQV LQSQLKWPVE FIYLGLRPAN NIAAGNTYQW RDWHHLTSVT NEPVY DVSQ SYARVSIDDT VAPVGSTTFK QSASQVMQNQ YIVPVETETL DTVRVKAHGI ELYAQYRAQF YRDYIPWNYG SFNLVT PQD KGALFLNFCL YPGTYQPSGH VNISRAREFY IEYTSSFCDS SNPCDLISIA KCINFLLISD GSAVLRYSTK EFYLQCL IL RCI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 / Details: Plunged into liquid ethane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient | ||||||||
| Output model |  PDB-5j7v: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)