[English] 日本語
 Yorodumi
Yorodumi- EMDB-7564: Mouse norovirus model using the crystal structure of MNV P domain... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7564 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse norovirus model using the crystal structure of MNV P domain and the Norwalkvirus shell domain | |||||||||
 Map data Map data | mouse norovirus (MNV-1) in PBS, pH 7.2 | |||||||||
 Sample Sample | mouse norovirus model != Murine norovirus 1 mouse norovirus model
| |||||||||
 Keywords Keywords | norovirus / mouse / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationantigenic variation / T=3 icosahedral viral capsid / symbiont-mediated perturbation of host defense response / virion component / host cell cytoplasm / identical protein binding Similarity search - Function | |||||||||
| Biological species |   Murine norovirus 1 Murine norovirus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||
 Authors Authors | Smith TJ | |||||||||
 Citation Citation | Journal: J Virol / Year: 2010 Title: High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. Authors: Umesh Katpally / Neil R Voss / Tommaso Cavazza / Stefan Taube / John R Rubin / Vivienne L Young / Jeanne Stuckey / Vernon K Ward / Herbert W Virgin / Christiane E Wobus / Thomas J Smith /  Abstract: Our previous structural studies on intact, infectious murine norovirus 1 (MNV-1) virions demonstrated that the receptor binding protruding (P) domains are lifted off the inner shell of the virus. ...Our previous structural studies on intact, infectious murine norovirus 1 (MNV-1) virions demonstrated that the receptor binding protruding (P) domains are lifted off the inner shell of the virus. Here, the three-dimensional (3D) reconstructions of recombinant rabbit hemorrhagic disease virus (rRHDV) virus-like particles (VLPs) and intact MNV-1 were determined to approximately 8-A resolution. rRHDV also has a raised P domain, and therefore, this conformation is independent of infectivity and genus. The atomic structure of the MNV-1 P domain was used to interpret the MNV-1 reconstruction. Connections between the P and shell domains and between the floating P domains were modeled. This observed P-domain flexibility likely facilitates virus-host receptor interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7564.map.gz emd_7564.map.gz | 169.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7564-v30.xml emd-7564-v30.xml emd-7564.xml emd-7564.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7564.png emd_7564.png | 325 KB | ||
| Filedesc metadata |  emd-7564.cif.gz emd-7564.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7564 http://ftp.pdbj.org/pub/emdb/structures/EMD-7564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7564 | HTTPS FTP |
-Related structure data
| Related structure data |  6crjMC  6c6qC  6c74C  6e47C  6e48C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7564.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7564.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mouse norovirus (MNV-1) in PBS, pH 7.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5599 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
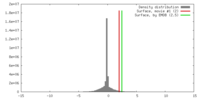
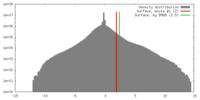
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : mouse norovirus model
| Entire | Name: mouse norovirus model |
|---|---|
| Components |
|
-Supramolecule #1: Murine norovirus 1
| Supramolecule | Name: Murine norovirus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: chimera of Norwalk virus shell domain and MNV P domain NCBI-ID: 223997 / Sci species name: Murine norovirus 1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: Norwalk virus, MNV-1 capsid protein chimera
| Macromolecule | Name: Norwalk virus, MNV-1 capsid protein chimera / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Murine norovirus 1 Murine norovirus 1 |
| Molecular weight | Theoretical: 57.127523 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SSVDGASGAG QLVPEVNASD PLAMDPVAGS STAVATAGQV NPIDPWIINN FVQAPQGEFT ISPNNTPGDV LFDLSLGPHL NPFLLHLSQ MYNGWVGNMR VRIMLAGNAF TAGKIIVSCI PPGFGSHNLT IAQATLFPHV IADVRTLDPI EVPLEDVRNV L FHNNDRNQ ...String: SSVDGASGAG QLVPEVNASD PLAMDPVAGS STAVATAGQV NPIDPWIINN FVQAPQGEFT ISPNNTPGDV LFDLSLGPHL NPFLLHLSQ MYNGWVGNMR VRIMLAGNAF TAGKIIVSCI PPGFGSHNLT IAQATLFPHV IADVRTLDPI EVPLEDVRNV L FHNNDRNQ QTMRLVCMLY TPLRTGGGTG DSFVVAGRVM TCPSPDFNFL FLVPAAAAAA RMVDLPVIQP RLCTHARWPA PV YGLLVDP SLPSNPQWQN GRVHVDGTLL GTTPISGSWV SCFAAEAAYE FQSGTGEVAT FTLIEQDGSA YVPGDRAAPL GYP DFSGQL EIEVQTETTK TGDKLKVTTF EMILGPTTNA DQAPYQGRVF ASVTAAASLD LVDGRVRAVP RSIYGFQDTI PEYN DGLLV PLAPPIGPFL PGEVLLRFRT YMRQIDTADA AAEAIDCALP QEFVSWFASN AFTVQSEALL LRYRNTLTGQ LLFEC KLYN EGYIALSYSG SGPLTFPTDG IFEVVSWVPR LYQLASVGSL ATGRMLK UniProtKB: Capsid protein VP1, Capsid protein VP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 / Details: PBS |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (2k x 2k) / Average electron dose: 10.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 8.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 20425 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6crj: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)