+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7303 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of histone H3k4 methyltransferase | |||||||||
 Map data Map data | Structure of histone H3k4 methyltransferase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Histone H3K4 Methyltransferase / GENE REGULATION-Transferase complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of meiotic DNA double-strand break formation / Formation of WDR5-containing histone-modifying complexes / : / [histone H3]-lysine4 N-trimethyltransferase / histone H3K4 trimethyltransferase activity / Set1C/COMPASS complex / RMTs methylate histone arginines / : / subtelomeric heterochromatin formation / telomere maintenance ...regulation of meiotic DNA double-strand break formation / Formation of WDR5-containing histone-modifying complexes / : / [histone H3]-lysine4 N-trimethyltransferase / histone H3K4 trimethyltransferase activity / Set1C/COMPASS complex / RMTs methylate histone arginines / : / subtelomeric heterochromatin formation / telomere maintenance / chromosome / histone binding / methylation / chromosome, telomeric region / transcription cis-regulatory region binding / chromatin binding / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / positive regulation of transcription by RNA polymerase II / zinc ion binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |    | |||||||||
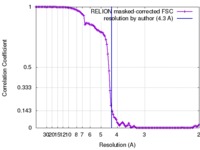
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Skiniotis G / Qu QH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex. Authors: Qianhui Qu / Yoh-Hei Takahashi / Yidai Yang / Hongli Hu / Yan Zhang / Joseph S Brunzelle / Jean-Francois Couture / Ali Shilatifard / Georgios Skiniotis /   Abstract: The methylation of histone 3 lysine 4 (H3K4) is carried out by an evolutionarily conserved family of methyltransferases referred to as complex of proteins associated with Set1 (COMPASS). The activity ...The methylation of histone 3 lysine 4 (H3K4) is carried out by an evolutionarily conserved family of methyltransferases referred to as complex of proteins associated with Set1 (COMPASS). The activity of the catalytic SET domain (su(var)3-9, enhancer-of-zeste, and trithorax) is endowed through forming a complex with a set of core proteins that are widely shared from yeast to humans. We obtained cryo-electron microscopy (cryo-EM) maps of the yeast Set1/COMPASS core complex at overall 4.0- to 4.4-Å resolution, providing insights into its structural organization and conformational dynamics. The Cps50 C-terminal tail weaves within the complex to provide a central scaffold for assembly. The SET domain, snugly positioned at the junction of the Y-shaped complex, is extensively contacted by Cps60 (Bre2), Cps50 (Swd1), and Cps30 (Swd3). The mobile SET-I motif of the SET domain is engaged by Cps30, explaining its key role in COMPASS catalytic activity toward higher H3K4 methylation states. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7303.map.gz emd_7303.map.gz | 107.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7303-v30.xml emd-7303-v30.xml emd-7303.xml emd-7303.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7303_fsc.xml emd_7303_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_7303.png emd_7303.png | 95.9 KB | ||
| Filedesc metadata |  emd-7303.cif.gz emd-7303.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7303 http://ftp.pdbj.org/pub/emdb/structures/EMD-7303 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7303 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7303 | HTTPS FTP |
-Validation report
| Summary document |  emd_7303_validation.pdf.gz emd_7303_validation.pdf.gz | 622.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7303_full_validation.pdf.gz emd_7303_full_validation.pdf.gz | 622.1 KB | Display | |
| Data in XML |  emd_7303_validation.xml.gz emd_7303_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_7303_validation.cif.gz emd_7303_validation.cif.gz | 15 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7303 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7303 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7303 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7303 | HTTPS FTP |
-Related structure data
| Related structure data |  6bx3MC  6e29C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7303.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7303.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of histone H3k4 methyltransferase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Multi-component complex of Set1 with its core subunits Cps25, Cps...
| Entire | Name: Multi-component complex of Set1 with its core subunits Cps25, Cps30, Cps40, Cps50 and Cps60 |
|---|---|
| Components |
|
-Supramolecule #1: Multi-component complex of Set1 with its core subunits Cps25, Cps...
| Supramolecule | Name: Multi-component complex of Set1 with its core subunits Cps25, Cps30, Cps40, Cps50 and Cps60 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 220 KDa |
-Macromolecule #1: Histone-lysine N-methyltransferase, H3 lysine-4 specific
| Macromolecule | Name: Histone-lysine N-methyltransferase, H3 lysine-4 specific type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: histone-lysine N-methyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.162402 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KIWQSRRKTL EEEKASDWQI ELNGTLFDSE LQPGSSFKAE GFRKVTDKLK INYLPHRRRV HQPLNTVNIH NERNEYTPEL CQREESSNK EPSDSVPQEV SSSRDNRASN RRFQQDIEAQ KAAIGTESEL LSLNQLNKRK KPVMFARSAI HNWGLYALDS I AAKEMIIE ...String: KIWQSRRKTL EEEKASDWQI ELNGTLFDSE LQPGSSFKAE GFRKVTDKLK INYLPHRRRV HQPLNTVNIH NERNEYTPEL CQREESSNK EPSDSVPQEV SSSRDNRASN RRFQQDIEAQ KAAIGTESEL LSLNQLNKRK KPVMFARSAI HNWGLYALDS I AAKEMIIE YVGERIRQPV AEMREKRYLK NGIGSSYLFR VDENTVIDAT KKGGIARFIN HCCDPNCTAK IIKVGGRRRI VI YALRDIA ASEELTYDYK FEREKDDEER LPCLCGAPNC K UniProtKB: Histone-lysine N-methyltransferase, H3 lysine-4 specific |
-Macromolecule #2: COMPASS component BRE2
| Macromolecule | Name: COMPASS component BRE2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 48.306922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FDHSGMSVMD RSEGLSISRD GNDLVSVPDQ YGWRTARSDV CIKEGMTYWE VEVIRGGNKK FADGVNNKEN ADDSVDEVQS GIYEKMHKQ VNDTPHLRFG VCRREASLEA PVGFDVYGYG IRDISLESIH EGKLNCVLEN GSPLKEGDKI GFLLSLPSIH T QIKQAKEF ...String: FDHSGMSVMD RSEGLSISRD GNDLVSVPDQ YGWRTARSDV CIKEGMTYWE VEVIRGGNKK FADGVNNKEN ADDSVDEVQS GIYEKMHKQ VNDTPHLRFG VCRREASLEA PVGFDVYGYG IRDISLESIH EGKLNCVLEN GSPLKEGDKI GFLLSLPSIH T QIKQAKEF TKRRIFALNS HMDTMNEPWR EDAENGPSRK KLKQETTNKE FQRALLEDIE YNDVVRDQIA IRYKNQLFFE AT DYVKTTK PEYYSSDKRE RQDYYQLEDS YLAIFQNGKY LGKAFENLKP LLPPFSELQY NEKFYLGYWQ HGEARDESND KNT TSAKKK KQQQKKKKGL ILRNKYVNNN KLGYYPTISC FNGGTARIIS EEDKLEYLDQ IRSAYCVDGN SKVNTLDTLY KEQI AEDIV WDIIDELEQI AL UniProtKB: COMPASS component BRE2 |
-Macromolecule #3: COMPASS component SDC1
| Macromolecule | Name: COMPASS component SDC1 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 4.810553 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TRKYLNTNVT PHLLAGMRLI AVQQPEDPLR VLGEYLIEQS NI UniProtKB: COMPASS component SDC1 |
-Macromolecule #4: COMPASS component SPP1
| Macromolecule | Name: COMPASS component SPP1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 28.189033 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HGREFVNDIW SRLKTDEDRA VVKKMVEQTG HIDKFKKFGQ LDFIDNNIVV KTDDEKEIFD QIVVRDMTLK TLEDDLQEVQ EISLPLFKK KLELLEVYLG WLDNVYTEMR KLDDDAASHV ECGKEDSKGT KRKKKKNSSR SRARKNICGY CSTYERIPCS V EEFVRDFG ...String: HGREFVNDIW SRLKTDEDRA VVKKMVEQTG HIDKFKKFGQ LDFIDNNIVV KTDDEKEIFD QIVVRDMTLK TLEDDLQEVQ EISLPLFKK KLELLEVYLG WLDNVYTEMR KLDDDAASHV ECGKEDSKGT KRKKKKNSSR SRARKNICGY CSTYERIPCS V EEFVRDFG SNEEATKIHE VCTKWKCNRH LDWVSTNQEQ YLQQIDSLES MQERLQHLIQ ARKKQLNIQY YEEILRRGL UniProtKB: COMPASS component SPP1 |
-Macromolecule #5: COMPASS component SWD1
| Macromolecule | Name: COMPASS component SWD1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 47.047637 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NILLQDPFAV LKEHPEKLTH TIENPLRTEC LQFSPCGDYL ALGCANGALV IYDMDTFRPI CVPGNMLGAH VRPITSIAWS PDGRLLLTS SRDWSIKLWD LSKPSKPLKE IRFDSPIWGC QWLDAKRRLC VATIFEESDA YVIDFSNDPV ASLLSKSDEK Q LSSTPDHG ...String: NILLQDPFAV LKEHPEKLTH TIENPLRTEC LQFSPCGDYL ALGCANGALV IYDMDTFRPI CVPGNMLGAH VRPITSIAWS PDGRLLLTS SRDWSIKLWD LSKPSKPLKE IRFDSPIWGC QWLDAKRRLC VATIFEESDA YVIDFSNDPV ASLLSKSDEK Q LSSTPDHG YVLVCTVHTK HPNIIIVGTS KGWLDFYKFH SLYQTECIHS LKITSSNIKH LIVSQNGERL AINCSDRTIR QY EISIDDE NSAVELTLEH KYQDVINKLQ WNCILFSNNT AEYLVASTHG SSAHELYIWE TTSGTLVRVL EGAEEELIDI NWD FYSMSI VSNGFESGNV YVWSVVIPPK WSALAPDFEE VEENVDYLEK EDEFDEVDEA EQQQGLEQEE EIAIDLRTRE QYDV RGNNL LVERFTI UniProtKB: COMPASS component SWD1 |
-Macromolecule #6: COMPASS component SWD3
| Macromolecule | Name: COMPASS component SWD3 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 34.655438 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FQFVTPVGTQ NGLKATCAKI SPDGQFLAIT QGLNILIYDI NRRTVSQTLV TSHARPFSEL CWSPDGQCIA TASDDFSVEI IHLSYGLLH TFIGHTAPVI SLTFNRKGNL LFTSSMDESI KIWDTLNGSL MKTISAHSEA VVSVDVPMND SSILSSGSYD G LIRIFDAE ...String: FQFVTPVGTQ NGLKATCAKI SPDGQFLAIT QGLNILIYDI NRRTVSQTLV TSHARPFSEL CWSPDGQCIA TASDDFSVEI IHLSYGLLH TFIGHTAPVI SLTFNRKGNL LFTSSMDESI KIWDTLNGSL MKTISAHSEA VVSVDVPMND SSILSSGSYD G LIRIFDAE TGHCLKTLTY DKDWKRENGV VPISQVKFSE NARYLLVKSL DGVVKIWDCI GGCVVRTFQV QPLEKGVLHH SC GMDFLNP EDGSTPLVIS GYENGDIYCW NSDTKSLLQL LDGSLYHHSS PVMSIHCFGN IMCSLALNGD CCLWRWV UniProtKB: COMPASS component SWD3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 9.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)