[English] 日本語
 Yorodumi
Yorodumi- EMDB-7097: Doubly PafE-capped 20S core particle in Mycobacterium tuberculosis -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7097 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Doubly PafE-capped 20S core particle in Mycobacterium tuberculosis | |||||||||
 Map data Map data | Doubly PafE-capped 20S CP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Protein degradation / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defenses / proteasome accessory complex / zymogen binding / positive regulation of proteasomal protein catabolic process / proteasome binding / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / threonine-type endopeptidase activity / proteasome core complex, alpha-subunit complex / proteasomal protein catabolic process ...symbiont-mediated perturbation of host defenses / proteasome accessory complex / zymogen binding / positive regulation of proteasomal protein catabolic process / proteasome binding / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / threonine-type endopeptidase activity / proteasome core complex, alpha-subunit complex / proteasomal protein catabolic process / regulation of proteasomal protein catabolic process / proteasome complex / proteolysis involved in protein catabolic process / peptidoglycan-based cell wall / protein homooligomerization / modification-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / extracellular region / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
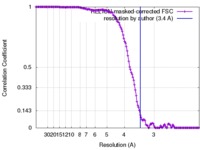
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Li H / Hu K | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2018 Journal: J Biol Chem / Year: 2018Title: Proteasome substrate capture and gate opening by the accessory factor PafE from . Authors: Kuan Hu / Jordan B Jastrab / Susan Zhang / Amanda Kovach / Gongpu Zhao / K Heran Darwin / Huilin Li /  Abstract: In all domains of life, proteasomes are gated, chambered proteases that require opening by activators to facilitate protein degradation. Twelve proteasome accessory factor E (PafE) monomers assemble ...In all domains of life, proteasomes are gated, chambered proteases that require opening by activators to facilitate protein degradation. Twelve proteasome accessory factor E (PafE) monomers assemble into a single dodecameric ring that promotes proteolysis required for the full virulence of the human bacterial pathogen Whereas the best characterized proteasome activators use ATP to deliver proteins into a proteasome, PafE does not require ATP. Here, to unravel the mechanism of PafE-mediated protein targeting and proteasome activation, we studied the interactions of PafE with native substrates, including a newly identified proteasome substrate, the ParA-like protein, Rv3213c, and with proteasome core particles. We characterized the function of a highly conserved feature in bacterial proteasome activator proteins: a glycine-glutamine-tyrosine-leucine (GQYL) motif at their C termini that is essential for stimulating proteolysis. Using cryo-electron microscopy (cryo-EM), we found that the GQYL motif of PafE interacts with specific residues in the α subunits of the proteasome core particle to trigger gate opening and degradation. Finally, we also found that PafE rings have 40-Å openings lined with hydrophobic residues that form a chamber for capturing substrates before they are degraded, suggesting PafE has a previously unrecognized chaperone activity. In summary, we have identified the interactions between PafE and the proteasome core particle that cause conformational changes leading to the opening of the proteasome gate and have uncovered a mechanism of PafE-mediated substrate degradation. Collectively, our results provide detailed insights into the mechanism of ATP-independent proteasome degradation in bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7097.map.gz emd_7097.map.gz | 193.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7097-v30.xml emd-7097-v30.xml emd-7097.xml emd-7097.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7097_fsc.xml emd_7097_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_7097.png emd_7097.png | 99.2 KB | ||
| Filedesc metadata |  emd-7097.cif.gz emd-7097.cif.gz | 5.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7097 http://ftp.pdbj.org/pub/emdb/structures/EMD-7097 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7097 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7097 | HTTPS FTP |
-Related structure data
| Related structure data |  6bglMC  7098C  6bgoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7097.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7097.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Doubly PafE-capped 20S CP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
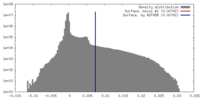
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Doubly PafE-capped 20S CP
| Entire | Name: Doubly PafE-capped 20S CP |
|---|---|
| Components |
|
-Supramolecule #1: Doubly PafE-capped 20S CP
| Supramolecule | Name: Doubly PafE-capped 20S CP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Proteasome subunit alpha
| Macromolecule | Name: Proteasome subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO / EC number: proteasome endopeptidase complex |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.911039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFPYFISPE QAMRERSELA RKGIARAKSV VALAYAGGVL FVAENPSRSL QKISELYDRV GFAAAGKFNE FDNLRRGGIQ FADTRGYAY DRRDVTGRQL ANVYAQTLGT IFTEQAKPYE VELCVAEVAH YGETKRPELY RITYDGSIAD EPHFVVMGGT T EPIANALK ...String: MSFPYFISPE QAMRERSELA RKGIARAKSV VALAYAGGVL FVAENPSRSL QKISELYDRV GFAAAGKFNE FDNLRRGGIQ FADTRGYAY DRRDVTGRQL ANVYAQTLGT IFTEQAKPYE VELCVAEVAH YGETKRPELY RITYDGSIAD EPHFVVMGGT T EPIANALK ESYAENASLT DALRIAVAAL RAGSADTSGG DQPTLGVASL EVAVLDANRP RRAFRRITGS ALQALLVDQE SP QSDGESS G UniProtKB: Proteasome subunit alpha |
-Macromolecule #2: Bacterial proteasome activator
| Macromolecule | Name: Bacterial proteasome activator / type: protein_or_peptide / ID: 2 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.963232 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVIGLSTGSD DDDVEVIGGV DPRLIAVQEN DSDESSLTDL VEQPAKVMRI GTMIKQLLEE VRAAPLDEAS RNRLRDIHAT SIRELEDGL APELREELDR LTLPFNEDAV PSDAELRIAQ AQLVGWLEGL FHGIQTALFA QQMAARAQLQ QMRQGALPPG V GKSGQHGH GTGQYL UniProtKB: Bacterial proteasome activator |
-Macromolecule #3: Proteasome subunit beta
| Macromolecule | Name: Proteasome subunit beta / type: protein_or_peptide / ID: 3 / Number of copies: 14 / Enantiomer: LEVO / EC number: proteasome endopeptidase complex |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.274264 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TTIVALKYPG GVVMAGDRRS TQGNMISGRD VRKVYITDDY TATGIAGTAA VAVEFARLYA VELEHYEKLE GVPLTFAGKI NRLAIMVRG NLAAAMQGLL ALPLLAGYDI HASDPQSAGR IVSFDAAGGW NIEEEGYQAV GSGSLFAKSS MKKLYSQVTD G DSGLRVAV ...String: TTIVALKYPG GVVMAGDRRS TQGNMISGRD VRKVYITDDY TATGIAGTAA VAVEFARLYA VELEHYEKLE GVPLTFAGKI NRLAIMVRG NLAAAMQGLL ALPLLAGYDI HASDPQSAGR IVSFDAAGGW NIEEEGYQAV GSGSLFAKSS MKKLYSQVTD G DSGLRVAV EALYDAADDD SATGGPDLVR GIFPTAVIID ADGAVDVPES RIAELARAII ESRSGADTFG SDGGEKHHHH HH UniProtKB: Proteasome subunit beta |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)