+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1C5H TCR bound to R-phycoerythrin | |||||||||
 Map data Map data | R-Phycoerythrin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | T-cell / gamma delta TCR / phycoerythrin / direct / IMMUNE SYSTEM | |||||||||
| Function / homology | Phycobilisome, alpha/beta subunit / Phycobilisome, alpha/beta subunit superfamily / Phycobilisome protein / phycobilisome / chloroplast thylakoid membrane / photosynthesis / Globin-like superfamily / R-phycoerythrin class I beta subunit / R-phycoerythrin class I alpha subunit Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Pyropia tenera (asakusa nori) Pyropia tenera (asakusa nori) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.68 Å | |||||||||
 Authors Authors | Rashleigh L / Venugopal H / Rossjohn J / Gully BS | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2025 Journal: Structure / Year: 2025Title: Antibody-like recognition of a γδ T cell receptor toward a foreign antigen. Authors: Liam Rashleigh / Hariprasad Venugopal / Michael T Rice / Sachith D Gunasinghe / Chhon Ling Sok / Nicholas A Gherardin / Catarina F Almeida / Ildiko Van Rhijn / D Branch Moody / Dale I ...Authors: Liam Rashleigh / Hariprasad Venugopal / Michael T Rice / Sachith D Gunasinghe / Chhon Ling Sok / Nicholas A Gherardin / Catarina F Almeida / Ildiko Van Rhijn / D Branch Moody / Dale I Godfrey / Jamie Rossjohn / Benjamin S Gully /    Abstract: The antigen recognition principles of B cells and αβ T cells have been well described compared to those of the γδ T cell. By way of their specificity conferring receptor (γδTCR), γδ T cells ...The antigen recognition principles of B cells and αβ T cells have been well described compared to those of the γδ T cell. By way of their specificity conferring receptor (γδTCR), γδ T cells can directly bind proteinaceous antigens. A known γδ T cell and B cell model antigen is phycoerythrin (PE), a light harvesting protein from rhodophytes and cyanobacteria. Here we probed human γδTCR reactivity to PE, in which a Vδ1Vγ5 TCR bound directly to induce proximal signaling and cellular activation. We determined the cryoelectron microscopy (cryo-EM) structure of the γδTCR-phycoerythrin immune complex. We then determined the cryo-EM structures of an antibody fragment and an αβTCR bound to PE. This revealed convergent use of apical aromatic residues to mediate contacts with a common PE epitope. Comparative analyses of the γδTCR revealed multiple antibody-like characteristics, including an enrichment of apical aromatic residues. Our findings reveal further distinct facets of antigen recognition by the γδTCR. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_70156.map.gz emd_70156.map.gz | 90.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-70156-v30.xml emd-70156-v30.xml emd-70156.xml emd-70156.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_70156_fsc.xml emd_70156_fsc.xml | 12.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_70156.png emd_70156.png | 109.3 KB | ||
| Masks |  emd_70156_msk_1.map emd_70156_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-70156.cif.gz emd-70156.cif.gz | 5.7 KB | ||
| Others |  emd_70156_additional_1.map.gz emd_70156_additional_1.map.gz emd_70156_half_map_1.map.gz emd_70156_half_map_1.map.gz emd_70156_half_map_2.map.gz emd_70156_half_map_2.map.gz | 167.7 MB 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-70156 http://ftp.pdbj.org/pub/emdb/structures/EMD-70156 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70156 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70156 | HTTPS FTP |
-Related structure data
| Related structure data |  9o61MC  9mgbC  9mkoC  9o60C  9o62C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_70156.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_70156.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | R-Phycoerythrin | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_70156_msk_1.map emd_70156_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: sharpened map
| File | emd_70156_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_70156_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_70156_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : R-phycoerythrin
| Entire | Name: R-phycoerythrin |
|---|---|
| Components |
|
-Supramolecule #1: R-phycoerythrin
| Supramolecule | Name: R-phycoerythrin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: R-phycoerythrin class I alpha subunit
| Macromolecule | Name: R-phycoerythrin class I alpha subunit / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pyropia tenera (asakusa nori) Pyropia tenera (asakusa nori) |
| Molecular weight | Theoretical: 17.707906 KDa |
| Sequence | String: MKSVITTTIS AADAAGRFPS SSDLESVQGN IQRAASRLEA AEKLAGNHEA VVKEAGDACF AKYPYLKNPG EAGDSQEKIN KCYRDIDHY MRLINYSLVV GGTGPLDEWG IAGAREVYRA LNLPGSSYIA AFVFTRDRLC VPRDMSAQAA VEFSGALDYV I NSLC UniProtKB: R-phycoerythrin class I alpha subunit |
-Macromolecule #2: R-phycoerythrin class I beta subunit
| Macromolecule | Name: R-phycoerythrin class I beta subunit / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pyropia tenera (asakusa nori) Pyropia tenera (asakusa nori) |
| Molecular weight | Theoretical: 18.33685 KDa |
| Sequence | String: MLDAFSRVVV NSDSKAAYVS GSDLQALKTF IADGNKRLDA VNSIVSNASC IVSDAVSGMI CENPGLIAPG GNCYTNRRMA ACLRDGEII LRYTSYALLA GDSSVLEDRC LNGLKETYIA LGVPTNSTAR AVSIMKSSAV AFISNTAPQR KMATAAGDCS A LSSEVASY CDKVSAAI UniProtKB: R-phycoerythrin class I beta subunit |
-Macromolecule #3: PHYCOERYTHROBILIN
| Macromolecule | Name: PHYCOERYTHROBILIN / type: ligand / ID: 3 / Number of copies: 24 / Formula: PEB |
|---|---|
| Molecular weight | Theoretical: 588.694 Da |
| Chemical component information |  ChemComp-PEB: |
-Macromolecule #4: PHYCOUROBILIN
| Macromolecule | Name: PHYCOUROBILIN / type: ligand / ID: 4 / Number of copies: 6 / Formula: PUB |
|---|---|
| Molecular weight | Theoretical: 590.71 Da |
| Chemical component information | 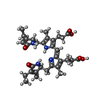 ChemComp-CYB: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















































