+Search query
-Structure paper
| Title | Antibody-like recognition of a γδ T cell receptor toward a foreign antigen. |
|---|---|
| Journal, issue, pages | Structure, Vol. 33, Issue 10, Page 1649-11662.e5, Year 2025 |
| Publish date | Oct 2, 2025 |
 Authors Authors | Liam Rashleigh / Hariprasad Venugopal / Michael T Rice / Sachith D Gunasinghe / Chhon Ling Sok / Nicholas A Gherardin / Catarina F Almeida / Ildiko Van Rhijn / D Branch Moody / Dale I Godfrey / Jamie Rossjohn / Benjamin S Gully /    |
| PubMed Abstract | The antigen recognition principles of B cells and αβ T cells have been well described compared to those of the γδ T cell. By way of their specificity conferring receptor (γδTCR), γδ T cells ...The antigen recognition principles of B cells and αβ T cells have been well described compared to those of the γδ T cell. By way of their specificity conferring receptor (γδTCR), γδ T cells can directly bind proteinaceous antigens. A known γδ T cell and B cell model antigen is phycoerythrin (PE), a light harvesting protein from rhodophytes and cyanobacteria. Here we probed human γδTCR reactivity to PE, in which a Vδ1Vγ5 TCR bound directly to induce proximal signaling and cellular activation. We determined the cryoelectron microscopy (cryo-EM) structure of the γδTCR-phycoerythrin immune complex. We then determined the cryo-EM structures of an antibody fragment and an αβTCR bound to PE. This revealed convergent use of apical aromatic residues to mediate contacts with a common PE epitope. Comparative analyses of the γδTCR revealed multiple antibody-like characteristics, including an enrichment of apical aromatic residues. Our findings reveal further distinct facets of antigen recognition by the γδTCR. |
 External links External links |  Structure / Structure /  PubMed:40744007 PubMed:40744007 |
| Methods | EM (single particle) |
| Resolution | 1.68 - 3.21 Å |
| Structure data | EMDB-48248, PDB-9mgb: EMDB-48332, PDB-9mko:  EMDB-70139: Phycoerythrin bound by 1C5H TCR (complex consensus map) EMDB-70155, PDB-9o60: EMDB-70156: 1C5H TCR bound to R-phycoerythrin EMDB-70157, PDB-9o62: |
| Chemicals |  ChemComp-PEB: 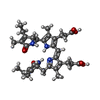 ChemComp-PUB: |
| Source |
|
 Keywords Keywords | IMMUNE SYSTEM / Phycoerythrin / antibody / scFv / immunity / TCR / direct recognition / T-cell / gamma delta TCR / direct |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













 neopyropia tenera (asakusa nori)
neopyropia tenera (asakusa nori)
 homo sapiens (human)
homo sapiens (human)