+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7007 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Mical Oxidized Actin (Class 1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | F-Actin / Mical / cytoskeleton / helix / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament ...cytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / skeletal muscle fiber development / stress fiber / titin binding / actin filament polymerization / actin filament / filopodium / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / lamellipodium / cell body / protein domain specific binding / hydrolase activity / calcium ion binding / positive regulation of gene expression / magnesium ion binding / ATP binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
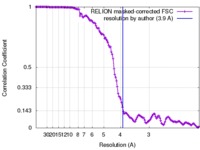
| Method | helical reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Grintsevich EE / Ge P | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Authors: Elena E Grintsevich / Peng Ge / Michael R Sawaya / Hunkar Gizem Yesilyurt / Jonathan R Terman / Z Hong Zhou / Emil Reisler /  Abstract: Actin filament assembly and disassembly are vital for cell functions. MICAL Redox enzymes are important post-translational effectors of actin that stereo-specifically oxidize actin's M44 and M47 ...Actin filament assembly and disassembly are vital for cell functions. MICAL Redox enzymes are important post-translational effectors of actin that stereo-specifically oxidize actin's M44 and M47 residues to induce cellular F-actin disassembly. Here we show that Mical-oxidized (Mox) actin can undergo extremely fast (84 subunits/s) disassembly, which depends on F-actin's nucleotide-bound state. Using near-atomic resolution cryoEM reconstruction and single filament TIRF microscopy we identify two dynamic and structural states of Mox-actin. Modeling actin's D-loop region based on our 3.9 Å cryoEM reconstruction suggests that oxidation by Mical reorients the side chain of M44 and induces a new intermolecular interaction of actin residue M47 (M47-O-T351). Site-directed mutagenesis reveals that this interaction promotes Mox-actin instability. Moreover, we find that Mical oxidation of actin allows for cofilin-mediated severing even in the presence of inorganic phosphate. Thus, in conjunction with cofilin, Mical oxidation of actin promotes F-actin disassembly independent of the nucleotide-bound state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7007.map.gz emd_7007.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7007-v30.xml emd-7007-v30.xml emd-7007.xml emd-7007.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7007_fsc.xml emd_7007_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_7007.png emd_7007.png | 52 KB | ||
| Filedesc metadata |  emd-7007.cif.gz emd-7007.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7007 http://ftp.pdbj.org/pub/emdb/structures/EMD-7007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7007 | HTTPS FTP |
-Related structure data
| Related structure data |  6av9MC  7008C  5uboC  6avbC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7007.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7007.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.041 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Filamentous Actin Oxidized by Mical at M44 and M47
| Entire | Name: Filamentous Actin Oxidized by Mical at M44 and M47 |
|---|---|
| Components |
|
-Supramolecule #1: Filamentous Actin Oxidized by Mical at M44 and M47
| Supramolecule | Name: Filamentous Actin Oxidized by Mical at M44 and M47 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.141973 KDa |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGV(SME)VG(SME)G QKDSYVGDEA QSKRGILTLK YP IE(HIC)GIIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASG RTTGIVLDSG ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGV(SME)VG(SME)G QKDSYVGDEA QSKRGILTLK YP IE(HIC)GIIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASG RTTGIVLDSG DGVTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSSS LEKSYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVMSGGTTM Y PGIADRMQ KEITALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWITKQEYDE AGPSIVHRKC F UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 240 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: Blot Force 1, Blot time 4s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: Gatan Bio Quantum |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 4-20 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6av9: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)