[English] 日本語
 Yorodumi
Yorodumi- EMDB-6945: Structure of human mitochondrial trifunctional protein, tetramer,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6945 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human mitochondrial trifunctional protein, tetramer, reconstruction with mask including detergent micelle applied | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Liang K / Li N / Dai J / Wang X / Liu P / Chen X / Wang C / Gao N / Xiao J | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Cryo-EM structure of human mitochondrial trifunctional protein. Authors: Kai Liang / Ningning Li / Xiao Wang / Jianye Dai / Pulan Liu / Chu Wang / Xiao-Wei Chen / Ning Gao / Junyu Xiao /  Abstract: The mitochondrial trifunctional protein (TFP) catalyzes three reactions in the fatty acid β-oxidation process. Mutations in the two TFP subunits cause mitochondrial trifunctional protein deficiency ...The mitochondrial trifunctional protein (TFP) catalyzes three reactions in the fatty acid β-oxidation process. Mutations in the two TFP subunits cause mitochondrial trifunctional protein deficiency and acute fatty liver of pregnancy that can lead to death. Here we report a 4.2-Å cryo-electron microscopy α2β2 tetrameric structure of the human TFP. The tetramer has a V-shaped architecture that displays a distinct assembly compared with the bacterial TFPs. A concave surface of the TFP tetramer interacts with the detergent molecules in the structure, suggesting that this region is involved in associating with the membrane. Deletion of a helical hairpin in TFPβ decreases its binding to the liposomes in vitro and reduces its membrane targeting in cells. Our results provide the structural basis for TFP function and have important implications for fatty acid oxidation related diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6945.map.gz emd_6945.map.gz | 20.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6945-v30.xml emd-6945-v30.xml emd-6945.xml emd-6945.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6945.png emd_6945.png | 184.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6945 http://ftp.pdbj.org/pub/emdb/structures/EMD-6945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6945 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6945.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6945.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
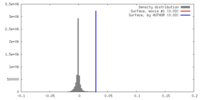
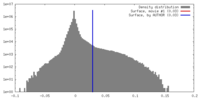
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human mitochondrial trifunctional protein
| Entire | Name: human mitochondrial trifunctional protein |
|---|---|
| Components |
|
-Supramolecule #1: human mitochondrial trifunctional protein
| Supramolecule | Name: human mitochondrial trifunctional protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.4 Å / Resolution method: FSC 0.143 CUT-OFF / Details: Gold Standard / Number images used: 426969 |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)