[English] 日本語
 Yorodumi
Yorodumi- EMDB-6888: Cryo-EM structure of HBV Cp164 capsid treated with micrococcal nu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6888 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HBV Cp164 capsid treated with micrococcal nuclease | |||||||||
 Map data Map data | HBV Cp-164 capsid treated with micrococcal nuclease | |||||||||
 Sample Sample | HBV core protein-164 != Hepatitis B virus HBV core protein-164
| |||||||||
| Biological species |   Hepatitis B virus Hepatitis B virus | |||||||||
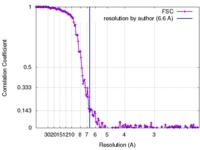
| Method | single particle reconstruction / cryo EM / Resolution: 6.6 Å | |||||||||
 Authors Authors | He J / Li Z / Gao Y / Yan D / Sun J / Zhang Q / Liu S | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Insights into the Structural Organization of Carboxy-Terminal Domain via Nuclease Sensitivity of Hepatitis B Virus Capsid Authors: He J / Li Z / Gao Y / Yan D / Sun J / Zhang Q / Liu S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6888.map.gz emd_6888.map.gz | 228.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6888-v30.xml emd-6888-v30.xml emd-6888.xml emd-6888.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6888_fsc.xml emd_6888_fsc.xml | 16 KB | Display |  FSC data file FSC data file |
| Images |  emd_6888.png emd_6888.png | 318.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6888 http://ftp.pdbj.org/pub/emdb/structures/EMD-6888 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6888 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6888 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6888.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6888.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HBV Cp-164 capsid treated with micrococcal nuclease | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HBV core protein-164
| Entire | Name: HBV core protein-164 |
|---|---|
| Components |
|
-Supramolecule #1: Hepatitis B virus
| Supramolecule | Name: Hepatitis B virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10407 / Sci species name: Hepatitis B virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host system | Organism:  |
| Virus shell | Shell ID: 1 / T number (triangulation number): 4 |
-Macromolecule #1: HBV core protein-164
| Macromolecule | Name: HBV core protein-164 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis B virus Hepatitis B virus |
| Recombinant expression | Organism:  |
| Sequence | String: MDIDPYKEFG ASVELLSFLP SDFFPSIRDL LDTASALYRE ALESPEHCSP HHTALRQAIL CWGELMNLAT WVGSNLEDPA SRELVVSYVN VNMGLKIRQL LWFHISCLTF GRETVLEYLV SFGVWIRTPP AYRPPNAPIL STLPETTVVR RRGRSPRRRT PSPR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)