+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6385 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of AP-1:Arf1:Nef complex | |||||||||
 Map data Map data | Trimeric structure of AP-1:Arf1:Nef | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adaptor protein-1 / Arf1 / Nef / clathrin cage | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.0 Å | |||||||||
 Authors Authors | Shen QT / Ren X / Zhang R / Lee H / Hurley J | |||||||||
 Citation Citation |  Journal: Science / Year: 2015 Journal: Science / Year: 2015Title: HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Authors: Qing-Tao Shen / Xuefeng Ren / Rui Zhang / Il-Hyung Lee / James H Hurley /  Abstract: The lentiviruses HIV and simian immunodeficiency virus (SIV) subvert intracellular membrane traffic as part of their replication cycle. The lentiviral Nef protein helps viruses evade innate and ...The lentiviruses HIV and simian immunodeficiency virus (SIV) subvert intracellular membrane traffic as part of their replication cycle. The lentiviral Nef protein helps viruses evade innate and adaptive immune defenses by hijacking the adaptor protein 1 (AP-1) and AP-2 clathrin adaptors. We found that HIV-1 Nef and the guanosine triphosphatase Arf1 induced trimerization and activation of AP-1. Here we report the cryo-electron microscopy structures of the Nef- and Arf1-bound AP-1 trimer in the active and inactive states. A central nucleus of three Arf1 molecules organizes the trimers. We combined the open trimer with a known dimer structure and thus predicted a hexagonal assembly with inner and outer faces that bind the membranes and clathrin, respectively. Hexagons were directly visualized and the model validated by reconstituting clathrin cage assembly. Arf1 and Nef thus play interconnected roles in allosteric activation, cargo recruitment, and coat assembly, revealing an unexpectedly intricate organization of the inner AP-1 layer of the clathrin coat. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6385.map.gz emd_6385.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6385-v30.xml emd-6385-v30.xml emd-6385.xml emd-6385.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6385.jpg emd_6385.jpg | 149.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6385 http://ftp.pdbj.org/pub/emdb/structures/EMD-6385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6385 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6385.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6385.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Trimeric structure of AP-1:Arf1:Nef | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.64 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
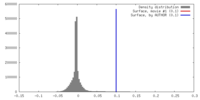
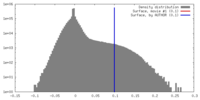
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AP-1:Arf1:Nef complex
| Entire | Name: AP-1:Arf1:Nef complex |
|---|---|
| Components |
|
-Supramolecule #1000: AP-1:Arf1:Nef complex
| Supramolecule | Name: AP-1:Arf1:Nef complex / type: sample / ID: 1000 / Oligomeric state: trimer / Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 850 KDa / Theoretical: 870 KDa / Method: multi-angle light scattering |
-Macromolecule #1: adaptor protein-1
| Macromolecule | Name: adaptor protein-1 / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: trans-Golgi network Homo sapiens (human) / synonym: Human / Location in cell: trans-Golgi network |
| Recombinant expression | Organism:  |
-Macromolecule #2: Arf1
| Macromolecule | Name: Arf1 / type: protein_or_peptide / ID: 2 / Name.synonym: ADP-ribosylation factor-1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human Homo sapiens (human) / synonym: human |
| Molecular weight | Experimental: 200 KDa / Theoretical: 200 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #3: Nef
| Macromolecule | Name: Nef / type: protein_or_peptide / ID: 3 / Name.synonym: negative factor / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human Homo sapiens (human) / synonym: human |
| Molecular weight | Experimental: 290 KDa / Theoretical: 290 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris, 200 mM NaCl, 0.3 mM TCEP |
| Grid | Details: 400 mesh copper grid with thin carbon support |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 120 K / Instrument: FEI VITROBOT MARK IV / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Date | Mar 30, 2015 |
| Image recording | Category: CCD / Film or detector model: DIRECT ELECTRON DE-12 (4k x 3k) / Number real images: 12000 / Average electron dose: 45.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 80000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | The particles were picked using the DoG Picker. |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.0 Å / Resolution method: OTHER / Software - Name: RELION / Number images used: 12000 |
| Final two d classification | Number classes: 10 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)