[English] 日本語
 Yorodumi
Yorodumi- EMDB-4025: Human Anaphase-promoting complex/Cyclosome (APC/C) APC15 deletion... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4025 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Anaphase-promoting complex/Cyclosome (APC/C) APC15 deletion mutant bound to the E2 UBE2C (aka UBCH10) poised for ubiquitin ligation to substrate. | |||||||||
 Map data Map data | Human Anaphase-promoting complex/Cyclosome (APC/C) APC15 deletion mutant bound to the E2 UBE2C (aka UBCH10) poised for ubiquitin ligation to substrate | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of diacylglycerol biosynthetic process / protein localization to septin ring / mitotic morphogenesis checkpoint signaling / cellular bud neck septin ring / metaphase/anaphase transition of cell cycle / metaphase/anaphase transition of meiosis I / Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components / mitotic checkpoint complex / positive regulation of anaphase-promoting complex-dependent catabolic process / positive regulation of exit from mitosis ...negative regulation of diacylglycerol biosynthetic process / protein localization to septin ring / mitotic morphogenesis checkpoint signaling / cellular bud neck septin ring / metaphase/anaphase transition of cell cycle / metaphase/anaphase transition of meiosis I / Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components / mitotic checkpoint complex / positive regulation of anaphase-promoting complex-dependent catabolic process / positive regulation of exit from mitosis / free ubiquitin chain polymerization / positive regulation of synapse maturation / regulation of meiotic nuclear division / Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase / regulation of dendrite development / Inactivation of APC/C via direct inhibition of the APC/C complex / APC/C:Cdc20 mediated degradation of mitotic proteins / positive regulation of synaptic plasticity / anaphase-promoting complex / Phosphorylation of Emi1 / Aberrant regulation of mitotic exit in cancer due to RB1 defects / regulation of meiotic cell cycle / anaphase-promoting complex-dependent catabolic process / metaphase/anaphase transition of mitotic cell cycle / protein branched polyubiquitination / Phosphorylation of the APC/C / anaphase-promoting complex binding / regulation of exit from mitosis / positive regulation of dendrite morphogenesis / cellular bud neck / positive regulation of mitotic metaphase/anaphase transition / (E3-independent) E2 ubiquitin-conjugating enzyme / positive regulation of ubiquitin protein ligase activity / exit from mitosis / ubiquitin ligase activator activity / protein K11-linked ubiquitination / regulation of mitotic metaphase/anaphase transition / mitotic sister chromatid cohesion / ubiquitin-ubiquitin ligase activity / E2 ubiquitin-conjugating enzyme / mitotic metaphase chromosome alignment / mitotic spindle assembly checkpoint signaling / Regulation of APC/C activators between G1/S and early anaphase / ubiquitin conjugating enzyme activity / ubiquitin-like protein ligase binding / Transcriptional Regulation by VENTX / cullin family protein binding / mitotic spindle assembly / enzyme-substrate adaptor activity / ubiquitin ligase complex / positive regulation of axon extension / ubiquitin-like ligase-substrate adaptor activity / heterochromatin / protein K48-linked ubiquitination / intercellular bridge / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Mitotic Prometaphase / nuclear periphery / APC/C:Cdc20 mediated degradation of Cyclin B / EML4 and NUDC in mitotic spindle formation / regulation of mitotic cell cycle / APC-Cdc20 mediated degradation of Nek2A / Resolution of Sister Chromatid Cohesion / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / SCF-beta-TrCP mediated degradation of Emi1 / Assembly of the pre-replicative complex / protein catabolic process / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / RHO GTPases Activate Formins / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / G protein-coupled receptor binding / brain development / kinetochore / CDK-mediated phosphorylation and removal of Cdc6 / G2/M transition of mitotic cell cycle / histone deacetylase binding / spindle / neuron projection development / protein polyubiquitination / spindle pole / ubiquitin-protein transferase activity / mitotic spindle / ubiquitin protein ligase activity / Separation of Sister Chromatids / nervous system development / mitotic cell cycle / Antigen processing: Ubiquitination & Proteasome degradation / microtubule cytoskeleton / Senescence-Associated Secretory Phenotype (SASP) / ubiquitin-dependent protein catabolic process / protein phosphatase binding / molecular adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / cell differentiation / protein kinase activity / non-specific serine/threonine protein kinase / regulation of cell cycle / Ub-specific processing proteases Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.1 Å | |||||||||
 Authors Authors | Stark H / Schulman BA / Peters JM | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2016 Journal: Mol Cell / Year: 2016Title: Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation. Authors: Masaya Yamaguchi / Ryan VanderLinden / Florian Weissmann / Renping Qiao / Prakash Dube / Nicholas G Brown / David Haselbach / Wei Zhang / Sachdev S Sidhu / Jan-Michael Peters / Holger Stark ...Authors: Masaya Yamaguchi / Ryan VanderLinden / Florian Weissmann / Renping Qiao / Prakash Dube / Nicholas G Brown / David Haselbach / Wei Zhang / Sachdev S Sidhu / Jan-Michael Peters / Holger Stark / Brenda A Schulman /     Abstract: The mitotic checkpoint complex (MCC) coordinates proper chromosome biorientation on the spindle with ubiquitination activities of CDC20-activated anaphase-promoting complex/cyclosome (APC/C(CDC20)). ...The mitotic checkpoint complex (MCC) coordinates proper chromosome biorientation on the spindle with ubiquitination activities of CDC20-activated anaphase-promoting complex/cyclosome (APC/C(CDC20)). APC/C(CDC20) and two E2s, UBE2C and UBE2S, catalyze ubiquitination through distinct architectures for linking ubiquitin (UB) to substrates and elongating polyUB chains, respectively. MCC, which contains a second molecule of CDC20, blocks APC/C(CDC20)-UBE2C-dependent ubiquitination of Securin and Cyclins, while differentially determining or inhibiting CDC20 ubiquitination to regulate spindle surveillance, checkpoint activation, and checkpoint termination. Here electron microscopy reveals conformational variation of APC/C(CDC20)-MCC underlying this multifaceted regulation. MCC binds APC/C-bound CDC20 to inhibit substrate access. However, rotation about the CDC20-MCC assembly and conformational variability of APC/C modulate UBE2C-catalyzed ubiquitination of MCC's CDC20 molecule. Access of UBE2C is limiting for subsequent polyubiquitination by UBE2S. We propose that conformational dynamics of APC/C(CDC20)-MCC modulate E2 activation and determine distinctive ubiquitination activities as part of a response mechanism ensuring accurate sister chromatid segregation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4025.map.gz emd_4025.map.gz | 58.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4025-v30.xml emd-4025-v30.xml emd-4025.xml emd-4025.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4025.png emd_4025.png | 159.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4025 http://ftp.pdbj.org/pub/emdb/structures/EMD-4025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4025 | HTTPS FTP |
-Related structure data
| Related structure data |  5khrMC  4021C  4022C  4023C  4024C  4026C  4027C  4028C  5khuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4025.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4025.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human Anaphase-promoting complex/Cyclosome (APC/C) APC15 deletion mutant bound to the E2 UBE2C (aka UBCH10) poised for ubiquitin ligation to substrate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.57 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
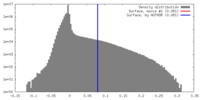
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human Anaphase-promoting complex/Cyclosome (APC/C) APC15 deletion...
| Entire | Name: Human Anaphase-promoting complex/Cyclosome (APC/C) APC15 deletion mutant bound to the E2 UBE2C (aka UBCH10) poised for ubiquitin ligation to substrate. |
|---|---|
| Components |
|
-Supramolecule #1: Human Anaphase-promoting complex/Cyclosome (APC/C) APC15 deletion...
| Supramolecule | Name: Human Anaphase-promoting complex/Cyclosome (APC/C) APC15 deletion mutant bound to the E2 UBE2C (aka UBCH10) poised for ubiquitin ligation to substrate. type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) / Recombinant plasmid: biGBac Trichoplusia ni (cabbage looper) / Recombinant plasmid: biGBac |
| Molecular weight | Theoretical: 1.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Grafix treated complex |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 1476 / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 0.0035 µm / Calibrated defocus min: 0.0007 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal magnification: 94000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)