[English] 日本語
 Yorodumi
Yorodumi- EMDB-61847: CryoEM structure of M. tuberculosis ClpP1P2 bound to bortezomib -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of M. tuberculosis ClpP1P2 bound to bortezomib | ||||||||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | Mycobacterium tuberculosis / Caseinolytic protease system / Activation / HYDROLASE | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationendopeptidase Clp / endopeptidase Clp complex / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / peptidoglycan-based cell wall / ATPase binding / serine-type endopeptidase activity / plasma membrane / cytosol Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |   Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | ||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.56 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Zhou B / Zhao H / Gao Y / Chen W / Zhang T / He J / Xiong X | ||||||||||||||||||||||||||||||
| Funding support |  China, 9 items China, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib Authors: Zhou B / Zhao H / Gao Y / Chen X / Zhang T / He J / Xiong X | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61847.map.gz emd_61847.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61847-v30.xml emd-61847-v30.xml emd-61847.xml emd-61847.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61847_fsc.xml emd_61847_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_61847.png emd_61847.png | 39.4 KB | ||
| Masks |  emd_61847_msk_1.map emd_61847_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-61847.cif.gz emd-61847.cif.gz | 5.9 KB | ||
| Others |  emd_61847_half_map_1.map.gz emd_61847_half_map_1.map.gz emd_61847_half_map_2.map.gz emd_61847_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61847 http://ftp.pdbj.org/pub/emdb/structures/EMD-61847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61847 | HTTPS FTP |
-Related structure data
| Related structure data |  9jvzMC  8ycxC  8yd0C  8yd1C  8yd2C  8yd4C  9jvpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_61847.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61847.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_61847_msk_1.map emd_61847_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61847_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61847_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of M. tuberculosis BTZ-ClpP1P2 complex
| Entire | Name: CryoEM structure of M. tuberculosis BTZ-ClpP1P2 complex |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of M. tuberculosis BTZ-ClpP1P2 complex
| Supramolecule | Name: CryoEM structure of M. tuberculosis BTZ-ClpP1P2 complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-dependent Clp protease proteolytic subunit 1
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv |
| Molecular weight | Theoretical: 19.383111 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SLTDSVYERL LSERIIFLGS EVNDEIANRL CAQILLLAAE DASKDISLYI NSPGGSISAG MAIYDTMVLA PCDIATYAMG MAASMGEFL LAAGTKGKRY ALPHARILMH QPLGGVTGSA ADIAIQAEQF AVIKKEMFRL NAEFTGQPIE RIEADSDRDR W FTAAEALE YGFVDHIITR UniProtKB: ATP-dependent Clp protease proteolytic subunit 1 |
-Macromolecule #2: ATP-dependent Clp protease proteolytic subunit 2
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv |
| Molecular weight | Theoretical: 21.43041 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LPSFIEHSSF GVKESNPYNK LFEERIIFLG VQVDDASAND IMAQLLVLES LDPDRDITMY INSPGGGFTS LMAIYDTMQY VRADIQTVC LGQAASAAAV LLAAGTPGKR MALPNARVLI HQPSLSGVIQ GQFSDLEIQA AEIERMRTLM ETTLARHTGK D AGVIRKDT ...String: LPSFIEHSSF GVKESNPYNK LFEERIIFLG VQVDDASAND IMAQLLVLES LDPDRDITMY INSPGGGFTS LMAIYDTMQY VRADIQTVC LGQAASAAAV LLAAGTPGKR MALPNARVLI HQPSLSGVIQ GQFSDLEIQA AEIERMRTLM ETTLARHTGK D AGVIRKDT DRDKILTAEE AKDYGIIDTV LEYRKLS UniProtKB: ATP-dependent Clp protease proteolytic subunit 2 |
-Macromolecule #3: N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL...
| Macromolecule | Name: N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE type: ligand / ID: 3 / Number of copies: 14 / Formula: BO2 |
|---|---|
| Molecular weight | Theoretical: 384.237 Da |
| Chemical component information | 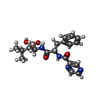 ChemComp-BO2: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)














































