[English] 日本語
 Yorodumi
Yorodumi- EMDB-61842: CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 3 | ||||||||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | Mycobacterium tuberculosis / Caseinolytic protease system / Activation / HYDROLASE | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to 11-deoxycorticosterone / response to dehydroepiandrosterone / negative regulation of lactation / potassium channel inhibitor activity / endopeptidase Clp / endopeptidase Clp complex / ATP-dependent peptidase activity / antioxidant activity / protein quality control for misfolded or incompletely synthesized proteins / cysteine-type endopeptidase inhibitor activity ...response to 11-deoxycorticosterone / response to dehydroepiandrosterone / negative regulation of lactation / potassium channel inhibitor activity / endopeptidase Clp / endopeptidase Clp complex / ATP-dependent peptidase activity / antioxidant activity / protein quality control for misfolded or incompletely synthesized proteins / cysteine-type endopeptidase inhibitor activity / negative regulation of tumor necrosis factor-mediated signaling pathway / negative regulation of canonical NF-kappaB signal transduction / protein folding chaperone / response to progesterone / peptidoglycan-based cell wall / Golgi lumen / negative regulation of inflammatory response / regulation of blood pressure / response to estradiol / ATPase binding / response to heat / serine-type endopeptidase activity / Golgi apparatus / protein homodimerization activity / ATP hydrolysis activity / extracellular space / ATP binding / metal ion binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |   Mycobacterium tuberculosis H37Rv (bacteria) / Mycobacterium tuberculosis H37Rv (bacteria) /  | ||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.15 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Zhou B / Zhao H / Gao Y / Chen X / Zhang T / He J / Xiong X | ||||||||||||||||||||||||||||||
| Funding support |  China, 9 items China, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib Authors: Zhou B / Zhao H / Gao Y / Chen X / Zhang T / He J / Xiong X | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61842.map.gz emd_61842.map.gz | 243.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61842-v30.xml emd-61842-v30.xml emd-61842.xml emd-61842.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61842_fsc.xml emd_61842_fsc.xml | 23.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_61842.png emd_61842.png | 35.5 KB | ||
| Masks |  emd_61842_msk_1.map emd_61842_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-61842.cif.gz emd-61842.cif.gz | 6.8 KB | ||
| Others |  emd_61842_half_map_1.map.gz emd_61842_half_map_1.map.gz emd_61842_half_map_2.map.gz emd_61842_half_map_2.map.gz | 471.1 MB 471.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61842 http://ftp.pdbj.org/pub/emdb/structures/EMD-61842 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61842 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61842 | HTTPS FTP |
-Validation report
| Summary document |  emd_61842_validation.pdf.gz emd_61842_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_61842_full_validation.pdf.gz emd_61842_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_61842_validation.xml.gz emd_61842_validation.xml.gz | 25.8 KB | Display | |
| Data in CIF |  emd_61842_validation.cif.gz emd_61842_validation.cif.gz | 34.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61842 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61842 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61842 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61842 | HTTPS FTP |
-Related structure data
| Related structure data |  9jvpMC  8ycxC  8yd0C  8yd1C  8yd2C  8yd4C  9jvzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61842.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61842.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_61842_msk_1.map emd_61842_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61842_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61842_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bo...
| Entire | Name: CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 3 |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bo...
| Supramolecule | Name: CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-dependent Clp protease ATP-binding subunit ClpC1
| Macromolecule | Name: ATP-dependent Clp protease ATP-binding subunit ClpC1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv |
| Molecular weight | Theoretical: 73.270336 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SLVLDQFGRN LTAAAMEGKL DPVIGREKEI ERVMQVLSRR TKNNPVLIGE PGVGKTAVVE GLAQAIVHGE VPETLKDKQL YTLDLGSLV AGSRYRGDFE ERLKKVLKEI NTRGDIILFI DALHTLVGAG AAEGAIDAAS ILKPKLARGE LQTIGATTLD E YRKYIEKD ...String: SLVLDQFGRN LTAAAMEGKL DPVIGREKEI ERVMQVLSRR TKNNPVLIGE PGVGKTAVVE GLAQAIVHGE VPETLKDKQL YTLDLGSLV AGSRYRGDFE ERLKKVLKEI NTRGDIILFI DALHTLVGAG AAEGAIDAAS ILKPKLARGE LQTIGATTLD E YRKYIEKD AALERRFQPV QVGEPTVEHT IEILKGLRDR YEAHHRVSIT DAAMVAAATL ADRYINDRFL PDKAIDLIDE AG ARMRIRR MTAPPDLREF DEKIAEARRE KESAIDAQDS EKAASLRDRE KTLVAQRAER EKQWRSGDLD VVAEVDDEQI AEV LGNWTG IPVFKLTEAE TTRLLRMEEE LHKRIIGQED AVKAVSKAIR RTRAGLKDPK RPSGSFIFAG PSGVGKTELS KALA NFLFG DDDALIQIDM GEFHDRFTAS RLFGAPPGYV GYEEGGQLTE KVRRKPFSVV LFDAIEKAHQ EIYNSLLQVL EDGRL TDGQ GRTVDFKNTV LIFTSNLGTS DISKPVGLGF SKGGGENDYE RMKQKVNDEL KKHFRPEFLN RIDDIIVFHQ LTREEI IRM VDLMISRVAG QLKSKDMALV LTDAAKALLA KRGFDPVLGA RPLRRTIQRE IEDQLSEKIL FEEVGPGQVV TVDVDNW DG EGPGEDAVFT FTGTR UniProtKB: ATP-dependent Clp protease ATP-binding subunit ClpC1 |
-Macromolecule #2: ATP-dependent Clp protease proteolytic subunit 2
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv |
| Molecular weight | Theoretical: 19.782576 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NPYNKLFEER IIFLGVQVDD ASANDIMAQL LVLESLDPDR DITMYINSPG GGFTSLMAIY DTMQYVRADI QTVCLGQAAS AAAVLLAAG TPGKRMALPN ARVLIHQPSL SGVIQGQFSD LEIQAAEIER MRTLMETTLA RHTGKDAGVI RKDTDRDKIL T AEEAKDYG IIDTVLEYRK LS UniProtKB: ATP-dependent Clp protease proteolytic subunit 2 |
-Macromolecule #3: ATP-dependent Clp protease proteolytic subunit 1
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit 1 / type: protein_or_peptide / ID: 3 / Number of copies: 7 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv |
| Molecular weight | Theoretical: 19.383111 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SLTDSVYERL LSERIIFLGS EVNDEIANRL CAQILLLAAE DASKDISLYI NSPGGSISAG MAIYDTMVLA PCDIATYAMG MAASMGEFL LAAGTKGKRY ALPHARILMH QPLGGVTGSA ADIAIQAEQF AVIKKEMFRL NAEFTGQPIE RIEADSDRDR W FTAAEALE YGFVDHIITR UniProtKB: ATP-dependent Clp protease proteolytic subunit 1 |
-Macromolecule #4: Beta-casein
| Macromolecule | Name: Beta-casein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.624252 KDa |
| Sequence | String: MKVLILACLV ALALARELEE LNVP UniProtKB: Beta-casein |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 8 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 8 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 4 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #8: N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL...
| Macromolecule | Name: N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE type: ligand / ID: 8 / Number of copies: 14 / Formula: BO2 |
|---|---|
| Molecular weight | Theoretical: 384.237 Da |
| Chemical component information | 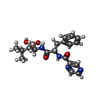 ChemComp-BO2: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)














































