+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | portal-tail of Vibrio cholerae typing phage release VP1 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | phage / virus / vibrio cholera phage / VIRAL PROTEIN | ||||||||||||
| Biological species |   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.7 Å | ||||||||||||
 Authors Authors | Liu HR / Pang H | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Three-dimensional structures of Vibrio cholerae typing podophage VP1 in two states. Authors: Hao Pang / Fenxia Fan / Jing Zheng / Hao Xiao / Zhixue Tan / Jingdong Song / Biao Kan / Hongrong Liu /  Abstract: Lytic podophages (VP1-VP5) play crucial roles in subtyping Vibrio cholerae O1 biotype El Tor. However, until now no structures of these phages have been available, which hindered our understanding of ...Lytic podophages (VP1-VP5) play crucial roles in subtyping Vibrio cholerae O1 biotype El Tor. However, until now no structures of these phages have been available, which hindered our understanding of the molecular mechanisms of infection and DNA release. Here, we determined the cryoelectron microscopy (cryo-EM) structures of mature and DNA-ejected VP1 structures at near-atomic and subnanometer resolutions, respectively. The VP1 head is composed of 415 copies of the major capsid protein gp7 and 11 turret-shaped spikes. The VP1 tail consists of an adapter, a nozzle, a slender ring, and a tail needle, and is flanked by three extended fibers I and six trimeric fibers II. Conformational changes of fiber II in DNA-ejected VP1 may cause the release of the tail needle and core proteins, forming an elongated tail channel. Our structures provide insights into the molecular mechanisms of infection and DNA release for podophages with a tail needle. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60199.map.gz emd_60199.map.gz | 226 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60199-v30.xml emd-60199-v30.xml emd-60199.xml emd-60199.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60199.png emd_60199.png | 65.9 KB | ||
| Filedesc metadata |  emd-60199.cif.gz emd-60199.cif.gz | 6.8 KB | ||
| Others |  emd_60199_half_map_1.map.gz emd_60199_half_map_1.map.gz emd_60199_half_map_2.map.gz emd_60199_half_map_2.map.gz | 226 MB 226 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60199 http://ftp.pdbj.org/pub/emdb/structures/EMD-60199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60199 | HTTPS FTP |
-Related structure data
| Related structure data |  8zkmMC  8zkkC  9in6C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60199.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60199.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
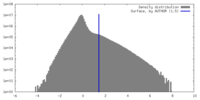
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60199_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
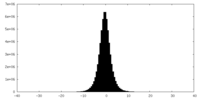
| Density Histograms |
-Half map: #1
| File | emd_60199_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
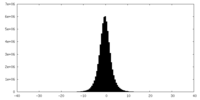
| Density Histograms |
- Sample components
Sample components
-Entire : Vibrio cholerae phage
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Vibrio cholerae phage
| Supramolecule | Name: Vibrio cholerae phage / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 666 / Sci species name: Vibrio cholerae phage / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: ring protein of release VP1
| Macromolecule | Name: ring protein of release VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.311119 KDa |
| Sequence | String: MSSWDRFDTP WDSIIDESTW EHYTFKYDAS FEAFSSMEVD EDLTITVSVL FASTSDNTVT VGQVLGLTMD NRAGSYFGAG SSWTSEAAV NNEAGSGFQS SHALQLSYSV EDGVISSFGS EAFCSFYNTV SFESSSDVQA AVNSLYNLDV LFSSGSGDSE Q HYVVFGET ...String: MSSWDRFDTP WDSIIDESTW EHYTFKYDAS FEAFSSMEVD EDLTITVSVL FASTSDNTVT VGQVLGLTMD NRAGSYFGAG SSWTSEAAV NNEAGSGFQS SHALQLSYSV EDGVISSFGS EAFCSFYNTV SFESSSDVQA AVNSLYNLDV LFSSGSGDSE Q HYVVFGET ASFESLAEHE TSSQYITHVE CMFNSVVEFE VKTYDWGRPV KPVGSDWSTD TPVVGVWVNE IYNGNKDWGE S |
-Macromolecule #2: adaptor of release VP1
| Macromolecule | Name: adaptor of release VP1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.59285 KDa |
| Sequence | String: MDKATLVKTI AYRMGNVKGQ DTAIDFELAL SIERLEGQEF VPWFLLSENN FFEGTAQENR IPVPRGFIRE YEEGSLYLRR VAGTGKCLI KKSQDQLLKY EGMTGEPSHY SLTNQYFRIY PVPQEDFKVE LLFYRKSSTL NVEDNPWYEY AAELLVAETI W AMLSARRD ...String: MDKATLVKTI AYRMGNVKGQ DTAIDFELAL SIERLEGQEF VPWFLLSENN FFEGTAQENR IPVPRGFIRE YEEGSLYLRR VAGTGKCLI KKSQDQLLKY EGMTGEPSHY SLTNQYFRIY PVPQEDFKVE LLFYRKSSTL NVEDNPWYEY AAELLVAETI W AMLSARRD KMADYWKSVA ADQMRRLTIL DAERRLANQE IFMG |
-Macromolecule #3: nozzle of release VP1
| Macromolecule | Name: nozzle of release VP1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 58.925699 KDa |
| Sequence | String: MEVTHKQFDL SGFLPDQAPD TLKSGAISRG FNVKPTILGW EKSGGFRETN TAPTNEKDFI FFWSPAIGDN RWFSGGDKTV QQVEGNVVS DVSRTGGYTA AAAARWNAVN FNGVLLMNNE LDSPQYLAAS GKLEDFPNLP SNVRFRTVAV YKNFILGLGV N FGSGFLDD ...String: MEVTHKQFDL SGFLPDQAPD TLKSGAISRG FNVKPTILGW EKSGGFRETN TAPTNEKDFI FFWSPAIGDN RWFSGGDKTV QQVEGNVVS DVSRTGGYTA AAAARWNAVN FNGVLLMNNE LDSPQYLAAS GKLEDFPNLP SNVRFRTVAV YKNFILGLGV N FGSGFLDD EIYWSHQADP GTMPPNWDYA NAASDSGRTP LPSEGYCVTS EELGSMNIVY KSDSIWTMQL IGGQWIFRFE NK FPGQGIL NKKSVVSFEG KHFVVTQKDI IVHDGYQVRS VADKRVRNFF FTDMNSDYFE RVFVVKDPRV AEVYVFYPSK NSV DGLCDR ALVWNWRDDV WSLLNLRPLK HAAYGYEITG VSITWDNFVG GWESTGLWQA DEDVAKYAPV LHYSFRDVPK LLAP TPQAL FIDEEIEAVW EREDIVIGSI SRDGVPYQDY ERNKSVSSIS FDVDTTEPFD VYIGYKGSLE DSVEWEFAGT VNPME DKRL FCLLTAGLFS MRIISKAQTF ILRSYKITYE FAGEMWA |
-Macromolecule #4: portal of release VP1
| Macromolecule | Name: portal of release VP1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.026672 KDa |
| Sequence | String: MEILYTGASE SLHSTILKAL LERIDLGDTF ISDNYTRWQA TERYYMMYKI PNKKDKAAIE KWNKGDTDFK SLVMPYSYAQ LMTAHAYMV NVFLNRDPIF QTDSLNGDGT ERELALESML QYQVKAGEME PSLLVWFMDA LRYGVGVLGD YWEEHVFHQT V FEVKKTRV ...String: MEILYTGASE SLHSTILKAL LERIDLGDTF ISDNYTRWQA TERYYMMYKI PNKKDKAAIE KWNKGDTDFK SLVMPYSYAQ LMTAHAYMV NVFLNRDPIF QTDSLNGDGT ERELALESML QYQVKAGEME PSLLVWFMDA LRYGVGVLGD YWEEHVFHQT V FEVKKTRV VKGYEGCKTF NVMVYDFIPD PRVALCKYQE GEFFGRRLDL NVLDLKKGAK FGKYFNVEHA EALVAASKEE MY RRDPSIG QQRSLKDSTM TPKGKQVGDI SCVEIFVRLV PKDWGLGDSE FPEMWVFTVA DKKYIVAAEP VNTLDDKFPF HIL ECEIDG YMNKSRGLLE ISAPMNDILT WLFDSHMYNK RQIMNNQFIG DPSALVVKDV ESKEPGKFIR LRPIISQLPV TDVT AQNIQ DVQVVERNMQ RIVGVNDDVA GQSSPSSRRS ATEFRGTTSF ASSRLANLAY FFSVTGFRSL AKSLIVKTQQ LYTVE MKVK VAGDNIKGAQ SIIVKPEDIS GQFDIMPVDG TLPVDRMAQA QFWMQIMSMV AGNPVLGAEY RLGDIFSYTA RLAGLK GID KMKIRILDDD QILALILA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)