[English] 日本語
 Yorodumi
Yorodumi- EMDB-5935: Cryo-EM structure of Dengue virus serotype 3 in complex with huma... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5935 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Dengue virus serotype 3 in complex with human antibody 5J7 Fab | |||||||||
 Map data Map data | Cryo-EM reconstruction of Dengue virus 3 in complex with human antibody 5J7 Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dengue virus / human antibody / neutralization | |||||||||
| Function / homology |  Function and homology information Function and homology informationflavivirin / host cell mitochondrion / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / ribonucleoside triphosphate phosphatase activity / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / channel activity ...flavivirin / host cell mitochondrion / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / ribonucleoside triphosphate phosphatase activity / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / clathrin-dependent endocytosis of virus by host cell / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / methyltransferase cap1 activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated activation of host autophagy / serine-type endopeptidase activity / RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / fusion of virus membrane with host endosome membrane / viral envelope / lipid binding / virion attachment to host cell / host cell nucleus / virion membrane / structural molecule activity / ATP hydrolysis activity / proteolysis / extracellular region / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Dengue virus 3 Dengue virus 3 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Fibriansah G / Tan JL / Smith SA / de Alwis R / Ng T-S / Kostyuchenko VA / Kukkaro P / de Silva AM / Crowe Jr JE / Lok S-M | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2015 Journal: Nat Commun / Year: 2015Title: A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Authors: Guntur Fibriansah / Joanne L Tan / Scott A Smith / Ruklanthi de Alwis / Thiam-Seng Ng / Victor A Kostyuchenko / Ramesh S Jadi / Petra Kukkaro / Aravinda M de Silva / James E Crowe / Shee-Mei Lok /   Abstract: Dengue virus (DENV) infects ~400 million people annually. There is no licensed vaccine or therapeutic drug. Only a small fraction of the total DENV-specific antibodies in a naturally occurring dengue ...Dengue virus (DENV) infects ~400 million people annually. There is no licensed vaccine or therapeutic drug. Only a small fraction of the total DENV-specific antibodies in a naturally occurring dengue infection consists of highly neutralizing antibodies. Here we show that the DENV-specific human monoclonal antibody 5J7 is exceptionally potent, neutralizing 50% of virus at nanogram-range antibody concentration. The 9 Å resolution cryo-electron microscopy structure of the Fab 5J7-DENV complex shows that a single Fab molecule binds across three envelope proteins and engages three functionally important domains, each from a different envelope protein. These domains are critical for receptor binding and fusion to the endosomal membrane. The ability to bind to multiple domains allows the antibody to fully coat the virus surface with only 60 copies of Fab, that is, half the amount compared with other potent antibodies. Our study reveals a highly efficient and unusual mechanism of molecular recognition by an antibody. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5935.map.gz emd_5935.map.gz | 290.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5935-v30.xml emd-5935-v30.xml emd-5935.xml emd-5935.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5935.png emd_5935.png | 394.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5935 http://ftp.pdbj.org/pub/emdb/structures/EMD-5935 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5935 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5935 | HTTPS FTP |
-Related structure data
| Related structure data |  3j6uMC  5933C  5934C  3j6sC  3j6tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5935.map.gz / Format: CCP4 / Size: 976.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5935.map.gz / Format: CCP4 / Size: 976.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of Dengue virus 3 in complex with human antibody 5J7 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
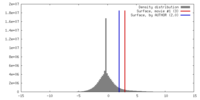
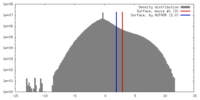
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dengue virus 3 complexed with human antibody 5J7 Fab
| Entire | Name: Dengue virus 3 complexed with human antibody 5J7 Fab |
|---|---|
| Components |
|
-Supramolecule #1000: Dengue virus 3 complexed with human antibody 5J7 Fab
| Supramolecule | Name: Dengue virus 3 complexed with human antibody 5J7 Fab / type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Supramolecule #1: Dengue virus 3
| Supramolecule | Name: Dengue virus 3 / type: virus / ID: 1 / NCBI-ID: 11069 / Sci species name: Dengue virus 3 / Sci species strain: D3/SG/05K863DK1/2005 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Host system | Organism:  |
-Macromolecule #1: antibody 5J7 Fab
| Macromolecule | Name: antibody 5J7 Fab / type: protein_or_peptide / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human Homo sapiens (human) / synonym: human |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 10 mM Tris-HCl, pH 8.0, 120 mM NaCl, 1 mM EDTA |
|---|---|
| Grid | Details: ultra-thin carbon-coated lacey carbon grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 100 K / Instrument: FEI VITROBOT MARK IV Method: Blotted with filter paper for 2 seconds prior to snap freezing |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Average: 100 K |
| Date | Mar 23, 2012 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON I (4k x 4k) / Digitization - Sampling interval: 14 µm / Number real images: 254 / Average electron dose: 18 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.2 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were manually selected. |
|---|---|
| CTF correction | Details: each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.0 Å / Resolution method: OTHER / Software - Name: MPSA, EMAN, EMAN2 / Number images used: 970 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D / Chain - #4 - Chain ID: E / Chain - #5 - Chain ID: F |
|---|---|
| Software | Name: Chimera, Coot, NAMD/MDFF |
| Details | Initially fitted in Chimera, model rebuilt in Coot, refined in NAMD/MDFF |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: real space correlation |
| Output model |  PDB-3j6u: |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera, Coot, NAMD/MDFF |
| Details | Initially fitted in Chimera, model rebuilt in Coot, refined in NAMD/MDFF |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: real space correlation |
| Output model |  PDB-3j6u: |
-Atomic model buiding 3
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera, Coot, NAMD/MDFF |
| Details | Initially fitted in Chimera, model rebuilt in Coot, refined in NAMD/MDFF |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: real space correlation |
| Output model |  PDB-3j6u: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)