+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5556 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain electron microscopy structure of Nup192 | |||||||||
 Map data Map data | Reconstruction of full-length Nup192 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nuclear Pore Complex / nuclear envelope / nuclear transport / nucleoporin | |||||||||
| Function / homology | nuclear pore inner ring / nuclear pore organization / structural constituent of nuclear pore / nucleocytoplasmic transport / nuclear pore Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Sampathkumar P / Kim SJ / Upla P / Rice W / Phillips J / Pieper U / Bonanno JB / Fernandez-Martinez J / Ketaren NE / Matsui T ...Sampathkumar P / Kim SJ / Upla P / Rice W / Phillips J / Pieper U / Bonanno JB / Fernandez-Martinez J / Ketaren NE / Matsui T / Stokes DL / Sauder JM / Burley SK / Sali A / Rout MP / Almo SC | |||||||||
 Citation Citation |  Journal: Structure / Year: 2013 Journal: Structure / Year: 2013Title: Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Authors: Parthasarathy Sampathkumar / Seung Joong Kim / Paula Upla / William J Rice / Jeremy Phillips / Benjamin L Timney / Ursula Pieper / Jeffrey B Bonanno / Javier Fernandez-Martinez / Zhanna ...Authors: Parthasarathy Sampathkumar / Seung Joong Kim / Paula Upla / William J Rice / Jeremy Phillips / Benjamin L Timney / Ursula Pieper / Jeffrey B Bonanno / Javier Fernandez-Martinez / Zhanna Hakhverdyan / Natalia E Ketaren / Tsutomu Matsui / Thomas M Weiss / David L Stokes / J Michael Sauder / Stephen K Burley / Andrej Sali / Michael P Rout / Steven C Almo /  Abstract: The nuclear pore complex, composed of proteins termed nucleoporins (Nups), is responsible for nucleocytoplasmic transport in eukaryotes. Nuclear pore complexes (NPCs) form an annular structure ...The nuclear pore complex, composed of proteins termed nucleoporins (Nups), is responsible for nucleocytoplasmic transport in eukaryotes. Nuclear pore complexes (NPCs) form an annular structure composed of the nuclear ring, cytoplasmic ring, a membrane ring, and two inner rings. Nup192 is a major component of the NPC's inner ring. We report the crystal structure of Saccharomyces cerevisiae Nup192 residues 2-960 [ScNup192(2-960)], which adopts an α-helical fold with three domains (i.e., D1, D2, and D3). Small angle X-ray scattering and electron microscopy (EM) studies reveal that ScNup192(2-960) could undergo long-range transition between "open" and "closed" conformations. We obtained a structural model of full-length ScNup192 based on EM, the structure of ScNup192(2-960), and homology modeling. Evolutionary analyses using the ScNup192(2-960) structure suggest that NPCs and vesicle-coating complexes are descended from a common membrane-coating ancestral complex. We show that suppression of Nup192 expression leads to compromised nuclear transport and hypothesize a role for Nup192 in modulating the permeability of the NPC central channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5556.map.gz emd_5556.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5556-v30.xml emd-5556-v30.xml emd-5556.xml emd-5556.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5556_1.png emd_5556_1.png | 30.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5556 http://ftp.pdbj.org/pub/emdb/structures/EMD-5556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5556 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5556.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5556.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of full-length Nup192 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.93 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
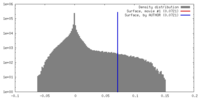
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Full-length Nup192
| Entire | Name: Full-length Nup192 |
|---|---|
| Components |
|
-Supramolecule #1000: Full-length Nup192
| Supramolecule | Name: Full-length Nup192 / type: sample / ID: 1000 Details: The sample was purified using affinity tags then centrifuged on a sucrose gradient. Fractions identified by SDS-PAGE and mass spectrometry. Oligomeric state: monomer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 192 KDa / Theoretical: 192 KDa / Method: SDS-PAGE |
-Macromolecule #1: Subunit of the Yeast Nuclear Pore Complex inner ring with weight ...
| Macromolecule | Name: Subunit of the Yeast Nuclear Pore Complex inner ring with weight 192 kDa type: protein_or_peptide / ID: 1 / Name.synonym: ScNup192 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 192 KDa / Theoretical: 192 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: structural constituent of nuclear pore, nuclear pore organization, nucleocytoplasmic transport, nuclear pore, nuclear pore inner ring |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.0035 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 20 mM HEPES, 300 mM NaCl, 2 mM MgCl2, 0.01% Tween 20, 0.01 mM DTT |
| Staining | Type: NEGATIVE Details: 3 ul protein was added to grids. The sample was blotted and 3 uL 1% uranyl formate added. Uranyl formate was blotted and fresh stain added (three times). The sample was allowed to sit 30 ...Details: 3 ul protein was added to grids. The sample was blotted and 3 uL 1% uranyl formate added. Uranyl formate was blotted and fresh stain added (three times). The sample was allowed to sit 30 seconds before final blot, then air-dried. |
| Grid | Details: 200 mesh copper grid with thin carbon support, glow discharged 20s with Gatan Solarus 20s in H2/O2. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | JEOL 2100F |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 250,000 times magnification |
| Details | Collected on 2028x2048 CCD, 24 um/pixel |
| Date | May 30, 2012 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F224 (2k x 2k) / Digitization - Sampling interval: 24 µm / Number real images: 400 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 81911 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: High tilt / Specimen holder model: JEOL / Tilt angle max: 50 |
- Electron microscopy #2
Electron microscopy #2
| Microscopy ID | 2 |
|---|---|
| Microscope | JEOL 2100F |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 250,000 times magnification |
| Details | Collected on 2028x2048 CCD, 24 um/pixel. 2 sessions are listed; data collected on multiple days between these dates |
| Date | Mar 1, 2012 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F224 (2k x 2k) / Digitization - Sampling interval: 24 µm / Number real images: 400 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 81911 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: High tilt / Specimen holder model: JEOL / Tilt angle max: 50 |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid Body. The domains were separately fitted by manual docking followed by "Fit in Map"using Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation 0.754 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)