[English] 日本語
 Yorodumi
Yorodumi- EMDB-5008: Paired beta-sheet structure of an Abeta(1-40) amyloid fibril reve... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5008 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Paired beta-sheet structure of an Abeta(1-40) amyloid fibril revealed by electron microscopy | |||||||||
 Map data Map data | The volume contains a total of 10 asymmetric units of the fibril | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Alzheimers disease / amyloid-beta / electron cryo-microscopy / protein folding / neurodegeneration | |||||||||
| Function / homology | Amyloidogenic glycoprotein, amyloid-beta peptide Function and homology information Function and homology information | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 8.8 Å | |||||||||
 Authors Authors | Sachse C / Faendrich M / Grigorieff N | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2008 Journal: Proc Natl Acad Sci U S A / Year: 2008Title: Paired beta-sheet structure of an Abeta(1-40) amyloid fibril revealed by electron microscopy. Authors: Carsten Sachse / Marcus Fändrich / Nikolaus Grigorieff /  Abstract: Alzheimer's disease is a neurodegenerative disorder that is characterized by the cerebral deposition of amyloid fibrils formed by Abeta peptide. Despite their prevalence in Alzheimer's and other ...Alzheimer's disease is a neurodegenerative disorder that is characterized by the cerebral deposition of amyloid fibrils formed by Abeta peptide. Despite their prevalence in Alzheimer's and other neurodegenerative diseases, important details of the structure of amyloid fibrils remain unknown. Here, we present a three-dimensional structure of a mature amyloid fibril formed by Abeta(1-40) peptide, determined by electron cryomicroscopy at approximately 8-A resolution. The fibril consists of two protofilaments, each containing approximately 5-nm-long regions of beta-sheet structure. A local twofold symmetry within each region suggests that pairs of beta-sheets are formed from equivalent parts of two Abeta(1-40) peptides contained in each protofilament. The pairing occurs via tightly packed interfaces, reminiscent of recently reported steric zipper structures. However, unlike these previous structures, the beta-sheet pairing is observed within an amyloid fibril and includes significantly longer amino acid sequences. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5008.map.gz emd_5008.map.gz | 2.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5008-v30.xml emd-5008-v30.xml emd-5008.xml emd-5008.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5008_1.tif emd_5008_1.tif | 164 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5008 http://ftp.pdbj.org/pub/emdb/structures/EMD-5008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5008 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5008.map.gz / Format: CCP4 / Size: 2.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5008.map.gz / Format: CCP4 / Size: 2.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The volume contains a total of 10 asymmetric units of the fibril | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human Abeta(1-40)
| Entire | Name: human Abeta(1-40) |
|---|---|
| Components |
|
-Supramolecule #1000: human Abeta(1-40)
| Supramolecule | Name: human Abeta(1-40) / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 4.33 KDa |
-Macromolecule #1: Abeta
| Macromolecule | Name: Abeta / type: protein_or_peptide / ID: 1 / Name.synonym: Abeta / Recombinant expression: No / Database: NCBI |
|---|---|
| Sequence | InterPro: Amyloidogenic glycoprotein, amyloid-beta peptide |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Details: 50 mM Borate |
| Grid | Details: Quantifoil R1.2/1.3 Cu 400 mesh grids |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 77.2 K / Instrument: OTHER / Details: Vitrification carried out in cold room at 277 K. / Method: Blot for 7 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 200,000 times magnification |
| Date | Nov 9, 2005 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 62 / Average electron dose: 35 e/Å2 / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 58333 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.9 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | 1. The fibrils in the sample were selected based on their uniform width. 2. A subset of limited crossover distances (130-150 nm) was chosen for the reconstruction. |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 4.8 Å Applied symmetry - Helical parameters - Δ&Phi: 1 ° Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER |
| CTF correction | Details: Each particle CTFTILT |
 Movie
Movie Controller
Controller



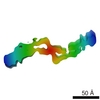










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)