[English] 日本語
 Yorodumi
Yorodumi- EMDB-11666: Nanodisc reconstituted, drug-free human ABCB1 in complex with MRK... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11666 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nanodisc reconstituted, drug-free human ABCB1 in complex with MRK16 Fab | |||||||||
 Map data Map data | drug-free human ABCB1-MRK16-Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | P-glycoprotein / MDR1 / nanodisc / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcarboxylic acid transmembrane transport / carboxylic acid transmembrane transporter activity / hormone transport / cellular response to nonylphenol / cellular response to borneol / response to codeine / response to cyclosporin A / cellular response to mycotoxin / daunorubicin transport / positive regulation of response to drug ...carboxylic acid transmembrane transport / carboxylic acid transmembrane transporter activity / hormone transport / cellular response to nonylphenol / cellular response to borneol / response to codeine / response to cyclosporin A / cellular response to mycotoxin / daunorubicin transport / positive regulation of response to drug / terpenoid transport / ceramide floppase activity / negative regulation of sensory perception of pain / positive regulation of establishment of Sertoli cell barrier / regulation of intestinal absorption / cellular response to external biotic stimulus / response to quercetin / response to antineoplastic agent / ceramide translocation / floppase activity / Abacavir transmembrane transport / establishment of blood-retinal barrier / protein localization to bicellular tight junction / phosphatidylethanolamine flippase activity / phosphatidylcholine floppase activity / external side of apical plasma membrane / Atorvastatin ADME / xenobiotic transport across blood-brain barrier / response to thyroxine / establishment of blood-brain barrier / transepithelial transport / xenobiotic detoxification by transmembrane export across the plasma membrane / export across plasma membrane / P-type phospholipid transporter / cellular response to L-glutamate / response to vitamin A / ABC-type xenobiotic transporter / response to vitamin D / response to glucagon / response to alcohol / intestinal absorption / response to glycoside / ABC-type xenobiotic transporter activity / Prednisone ADME / cellular response to antibiotic / phospholipid translocation / cellular hyperosmotic salinity response / maintenance of blood-brain barrier / cellular response to alkaloid / efflux transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / xenobiotic transmembrane transporter activity / transmembrane transporter activity / cellular response to dexamethasone stimulus / response to cadmium ion / transport across blood-brain barrier / lactation / xenobiotic metabolic process / regulation of chloride transport / response to progesterone / placenta development / stem cell proliferation / cellular response to estradiol stimulus / brush border membrane / female pregnancy / circadian rhythm / ABC-family proteins mediated transport / transmembrane transport / G2/M transition of mitotic cell cycle / cellular response to tumor necrosis factor / cellular response to lipopolysaccharide / response to hypoxia / apical plasma membrane / response to xenobiotic stimulus / ubiquitin protein ligase binding / cell surface / ATP hydrolysis activity / extracellular exosome / ATP binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
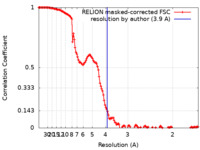
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Nosol K / Locher KP | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Cryo-EM structures reveal distinct mechanisms of inhibition of the human multidrug transporter ABCB1. Authors: Kamil Nosol / Ksenija Romane / Rossitza N Irobalieva / Amer Alam / Julia Kowal / Naoya Fujita / Kaspar P Locher /    Abstract: ABCB1 detoxifies cells by exporting diverse xenobiotic compounds, thereby limiting drug disposition and contributing to multidrug resistance in cancer cells. Multiple small-molecule inhibitors and ...ABCB1 detoxifies cells by exporting diverse xenobiotic compounds, thereby limiting drug disposition and contributing to multidrug resistance in cancer cells. Multiple small-molecule inhibitors and inhibitory antibodies have been developed for therapeutic applications, but the structural basis of their activity is insufficiently understood. We determined cryo-EM structures of nanodisc-reconstituted, human ABCB1 in complex with the Fab fragment of the inhibitory, monoclonal antibody MRK16 and bound to a substrate (the antitumor drug vincristine) or to the potent inhibitors elacridar, tariquidar, or zosuquidar. We found that inhibitors bound in pairs, with one molecule lodged in the central drug-binding pocket and a second extending into a phenylalanine-rich cavity that we termed the "access tunnel." This finding explains how inhibitors can act as substrates at low concentration, but interfere with the early steps of the peristaltic extrusion mechanism at higher concentration. Our structural data will also help the development of more potent and selective ABCB1 inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11666.map.gz emd_11666.map.gz | 202.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11666-v30.xml emd-11666-v30.xml emd-11666.xml emd-11666.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11666_fsc.xml emd_11666_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11666.png emd_11666.png | 108.8 KB | ||
| Filedesc metadata |  emd-11666.cif.gz emd-11666.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11666 http://ftp.pdbj.org/pub/emdb/structures/EMD-11666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11666 | HTTPS FTP |
-Related structure data
| Related structure data |  7a65MC  7a69C  7a6cC  7a6eC  7a6fC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11666.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11666.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | drug-free human ABCB1-MRK16-Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Nanodisc reconstituted, drug-free human ABCB1 in complex with MRK...
| Entire | Name: Nanodisc reconstituted, drug-free human ABCB1 in complex with MRK16 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Nanodisc reconstituted, drug-free human ABCB1 in complex with MRK...
| Supramolecule | Name: Nanodisc reconstituted, drug-free human ABCB1 in complex with MRK16 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 240 KDa |
-Supramolecule #2: Multidrug resistance protein 1
| Supramolecule | Name: Multidrug resistance protein 1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: MRK16 Fab-fragment light and heavy chain
| Supramolecule | Name: MRK16 Fab-fragment light and heavy chain / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Multidrug resistance protein 1
| Macromolecule | Name: Multidrug resistance protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ABC-type xenobiotic transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 141.628781 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDLEGDRNGG AKKKNFFKLN NKSEKDKKEK KPTVSVFSMF RYSNWLDKLY MVVGTLAAII HGAGLPLMML VFGEMTDIFA NAGNLEDLM SNITNRSDIN DTGFFMNLEE DMTRYAYYYS GIGAGVLVAA YIQVSFWCLA AGRQIHKIRK QFFHAIMRQE I GWFDVHDV ...String: MDLEGDRNGG AKKKNFFKLN NKSEKDKKEK KPTVSVFSMF RYSNWLDKLY MVVGTLAAII HGAGLPLMML VFGEMTDIFA NAGNLEDLM SNITNRSDIN DTGFFMNLEE DMTRYAYYYS GIGAGVLVAA YIQVSFWCLA AGRQIHKIRK QFFHAIMRQE I GWFDVHDV GELNTRLTDD VSKINEGIGD KIGMFFQSMA TFFTGFIVGF TRGWKLTLVI LAISPVLGLS AAVWAKILSS FT DKELLAY AKAGAVAEEV LAAIRTVIAF GGQKKELERY NKNLEEAKRI GIKKAITANI SIGAAFLLIY ASYALAFWYG TTL VLSGEY SIGQVLTVFF SVLIGAFSVG QASPSIEAFA NARGAAYEIF KIIDNKPSID SYSKSGHKPD NIKGNLEFRN VHFS YPSRK EVKILKGLNL KVQSGQTVAL VGNSGCGKST TVQLMQRLYD PTEGMVSVDG QDIRTINVRF LREIIGVVSQ EPVLF ATTI AENIRYGREN VTMDEIEKAV KEANAYDFIM KLPHKFDTLV GERGAQLSGG QKQRIAIARA LVRNPKILLL DEATSA LDT ESEAVVQVAL DKARKGRTTI VIAHRLSTVR NADVIAGFDD GVIVEKGNHD ELMKEKGIYF KLVTMQTAGN EVELENA AD ESKSEIDALE MSSNDSRSSL IRKRSTRRSV RGSQAQDRKL STKEALDESI PPVSFWRIMK LNLTEWPYFV VGVFCAII N GGLQPAFAII FSKIIGVFTR IDDPETKRQN SNLFSLLFLA LGIISFITFF LQGFTFGKAG EILTKRLRYM VFRSMLRQD VSWFDDPKNT TGALTTRLAN DAAQVKGAIG SRLAVITQNI ANLGTGIIIS FIYGWQLTLL LLAIVPIIAI AGVVEMKMLS GQALKDKKE LEGAGKIATE AIENFRTVVS LTQEQKFEHM YAQSLQVPYR NSLRKAHIFG ITFSFTQAMM YFSYAGCFRF G AYLVAHKL MSFEDVLLVF SAVVFGAMAV GQVSSFAPDY AKAKISAAHI IMIIEKTPLI DSYSTEGLMP NTLEGNVTFG EV VFNYPTR PDIPVLQGLS LEVKKGQTLA LVGSSGCGKS TVVQLLERFY DPLAGKVLLD GKEIKRLNVQ WLRAHLGIVS QEP ILFDCS IAENIAYGDN SRVVSQEEIV RAAKEANIHA FIESLPNKYS TKVGDKGTQL SGGQKQRIAI ARALVRQPHI LLLD EATSA LDTESEKVVQ EALDKAREGR TCIVIAHRLS TIQNADLIVV FQNGRVKEHG THQQLLAQKG IYFSMVSVQA GTKRQ UniProtKB: ATP-dependent translocase ABCB1 |
-Macromolecule #2: MRK16 Fab-fragment light chain
| Macromolecule | Name: MRK16 Fab-fragment light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.139758 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVLMTQTPVS LSVSLGDQAS ISCRSSQSIV HSTGNTYLEW YLQKPGQSPK LLIYKISNRF SGVPDRFSGS GSGTDFTLKI SRVEAEDLG VYYCFQASHA PRTFGGGTKL EIKRADAAPT VSIFPPSSEQ LTSGGASVVC FLNNFYPKDI NVKWKIDGSE R QNGVLNSW ...String: DVLMTQTPVS LSVSLGDQAS ISCRSSQSIV HSTGNTYLEW YLQKPGQSPK LLIYKISNRF SGVPDRFSGS GSGTDFTLKI SRVEAEDLG VYYCFQASHA PRTFGGGTKL EIKRADAAPT VSIFPPSSEQ LTSGGASVVC FLNNFYPKDI NVKWKIDGSE R QNGVLNSW TDQDSKDSTY SMSSTLTLTK DEYERHNSYT CEATHKTSTS PIVKSFNRNE C |
-Macromolecule #3: MRK16 Fab-fragment heavy chain
| Macromolecule | Name: MRK16 Fab-fragment heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.415236 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVILVESGGG LVKPGGSLKL SCAASGFTFS SYTMSWVRQT PEKRLEWVAT ISSGGGNTYY PDSVKGRFTI SRDNAKNNLY LQMSSLRSE DTALYYCARY YRYEAWFASW GQGTLVTVSA AKTTAPSVYP LAPVCGDTTG SSVTLGCLVK GYFPEPVTLT W NSGSLSSG ...String: EVILVESGGG LVKPGGSLKL SCAASGFTFS SYTMSWVRQT PEKRLEWVAT ISSGGGNTYY PDSVKGRFTI SRDNAKNNLY LQMSSLRSE DTALYYCARY YRYEAWFASW GQGTLVTVSA AKTTAPSVYP LAPVCGDTTG SSVTLGCLVK GYFPEPVTLT W NSGSLSSG VHTFPAVLQS DLYTLSSSVT VTSSTWPSQS ITCNVAHPAS STKVDKKIEP |
-Macromolecule #4: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 4 / Number of copies: 6 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)