+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SPEF1 bound to 13-PF microtubule | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | microtubule / spef1 / cilia / PROTEIN BINDING | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
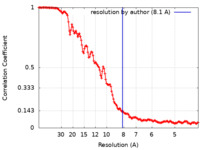
| Method | subtomogram averaging / cryo EM / Resolution: 8.1 Å | |||||||||
 Authors Authors | Legal T / Bui KH | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: Curr Biol / Year: 2025 Journal: Curr Biol / Year: 2025Title: Structure of the ciliary tip central pair reveals the unique role of the microtubule-seam binding protein SPEF1. Authors: Thibault Legal / Ewa Joachimiak / Mireya Parra / Wang Peng / Amanda Tam / Corbin Black / Mayukh Guha / Chau Anh Nguyen / Avrin Ghanaeian / Melissa Valente-Paterno / Gary Brouhard / Jacek ...Authors: Thibault Legal / Ewa Joachimiak / Mireya Parra / Wang Peng / Amanda Tam / Corbin Black / Mayukh Guha / Chau Anh Nguyen / Avrin Ghanaeian / Melissa Valente-Paterno / Gary Brouhard / Jacek Gaertig / Dorota Wloga / Khanh Huy Bui /    Abstract: Motile cilia are unique organelles with the ability to move autonomously. The force generated by beating cilia propels cells and moves fluids. The ciliary skeleton is made of peripheral doublet ...Motile cilia are unique organelles with the ability to move autonomously. The force generated by beating cilia propels cells and moves fluids. The ciliary skeleton is made of peripheral doublet microtubules and a central pair (CP) with a distinct structure at the tip. In this study, we present a high-resolution structure of the CP in the ciliary tip of the ciliate Tetrahymena thermophila and identify several tip proteins that bind and form unique patterns on both microtubules of the tip CP. Two of those proteins that contain tubulin polymerization-promoting protein (TPPP)-like domains, TLP1 and TLP2, bind to high curvature regions of the microtubule. TLP2, which contains two TPPP-like domains, is an unusually long protein that wraps laterally around half a microtubule and forms the bridge between the two microtubules. Moreover, we found that the conserved protein SPEF1 binds to both microtubule seams and crosslinked the two microtubules. In vitro, human SPEF1 binds to the microtubule seam as visualized by cryoelectron tomography and subtomogram averaging. Single-molecule microtubule dynamics assays indicate that SPEF1 stabilizes microtubules in vitro. Together, these data show that the proteins in the tip CP maintain stable microtubule structures and play important roles in maintaining the integrity of the axoneme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_49903.map.gz emd_49903.map.gz | 191.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-49903-v30.xml emd-49903-v30.xml emd-49903.xml emd-49903.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_49903_fsc.xml emd_49903_fsc.xml | 15.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_49903.png emd_49903.png | 340.8 KB | ||
| Filedesc metadata |  emd-49903.cif.gz emd-49903.cif.gz | 5.6 KB | ||
| Others |  emd_49903_half_map_1.map.gz emd_49903_half_map_1.map.gz emd_49903_half_map_2.map.gz emd_49903_half_map_2.map.gz | 105.9 MB 105.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-49903 http://ftp.pdbj.org/pub/emdb/structures/EMD-49903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-49903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-49903 | HTTPS FTP |
-Validation report
| Summary document |  emd_49903_validation.pdf.gz emd_49903_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_49903_full_validation.pdf.gz emd_49903_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_49903_validation.xml.gz emd_49903_validation.xml.gz | 20.4 KB | Display | |
| Data in CIF |  emd_49903_validation.cif.gz emd_49903_validation.cif.gz | 26.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-49903 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-49903 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-49903 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-49903 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_49903.map.gz / Format: CCP4 / Size: 206 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_49903.map.gz / Format: CCP4 / Size: 206 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.12 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_49903_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_49903_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : spef1 bound to a 13-pf microtubule
| Entire | Name: spef1 bound to a 13-pf microtubule |
|---|---|
| Components |
|
-Supramolecule #1: spef1 bound to a 13-pf microtubule
| Supramolecule | Name: spef1 bound to a 13-pf microtubule / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SPEF1
| Macromolecule | Name: SPEF1 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASSVDEEAL HQLYLWVDNI PLSRPKRNLS RDFSDGVLVA EVIKFYFPKM VEMHNYVPAN SLQQKLSNWG HLNRKVLKRL NFSVPDDVMR KIAQCAPGVV ELVLIPLRQR LEERQRRRKQ GAGSLQELAP QDGSGYMDVG VSQKARGEGV PDPQGGGQLS WDRPPAPRPP ...String: MASSVDEEAL HQLYLWVDNI PLSRPKRNLS RDFSDGVLVA EVIKFYFPKM VEMHNYVPAN SLQQKLSNWG HLNRKVLKRL NFSVPDDVMR KIAQCAPGVV ELVLIPLRQR LEERQRRRKQ GAGSLQELAP QDGSGYMDVG VSQKARGEGV PDPQGGGQLS WDRPPAPRPP AYNRALQGDP SFVLQIAEKE QELLASQETV QVLQMKVRRL EHLLQLKNVR IEDLSRRLQQ AERKQR |
| Recombinant expression | Organism:  |
-Macromolecule #2: tubulin alpha 1B
| Macromolecule | Name: tubulin alpha 1B / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS IYPAPQVSTA VVEPYNSILT THTTLEHSDC AFMVDNEAIY DICRRNLDIE RPTYTNLNRL ISQIVSSITA SLRFDGALNV DLTEFQTNLV PYPRIHFPLA TYAPVISAEK AYHEQLSVAE ITNACFEPAN QMVKCDPRHG KYMACCLLYR GDVVPKDVNA AIATIKTKRS IQFVDWCPTG FKVGINYQPP TVVPGGDLAK VQRAVCMLSN TTAIAEAWAR LDHKFDLMYA KRAFVHWYVG EGMEEGEFSE AREDMAALEK DYEEVGVDSV EGEGEEEGEE Y |
-Macromolecule #3: tubulin beta 2B
| Macromolecule | Name: tubulin beta 2B / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVM ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVM PSPKVSDTVV EPYNATLSVH QLVENTDETY CIDNEALYDI CFRTLKLTTP TYGDLNHLVS ATMSGVTTCL RFPGQLNADL RKLAVNMVPF PRLHFFMPGF APLTSRGSQQ YRALTVPELT QQMFDSKNMM AACDPRHGRY LTVAAIFRGR MSMKEVDEQM LNVQNKNSSY FVEWIPNNVK TAVCDIPPRG LKMSATFIGN STAIQELFKR ISEQFTAMFR RKAFLHWYTG EGMDEMEFTE AESNMNDLVS EYQQYQDATA DEQGEFEEEE GEDEA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 4.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)