+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4907 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molybdenum storage protein under turnover conditions | |||||||||
 Map data Map data | molybdenum storage protein under turnover conditions | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | molybdenum storage protein / ATPase / METAL BINDING PROTEIN | |||||||||
| Function / homology | Molybdenum storage protein subunit alpha/beta / nutrient reservoir activity / molybdenum ion binding / Aspartate/glutamate/uridylate kinase / Acetylglutamate kinase-like superfamily / Amino acid kinase family / cytoplasm / Molybdenum storage protein subunit beta / Molybdenum storage protein subunit alpha Function and homology information Function and homology information | |||||||||
| Biological species |  Azotobacter vinelandii DJ (bacteria) / Azotobacter vinelandii DJ (bacteria) /  Azotobacter vinelandii (strain DJ / ATCC BAA-1303) (bacteria) Azotobacter vinelandii (strain DJ / ATCC BAA-1303) (bacteria) | |||||||||
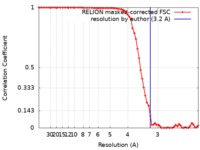
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Bruenle S / Mills DJ | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism. Authors: Steffen Brünle / Martin L Eisinger / Juliane Poppe / Deryck J Mills / Julian D Langer / Janet Vonck / Ulrich Ermler /  Abstract: The molybdenum storage protein (MoSto) deposits large amounts of molybdenum as polyoxomolybdate clusters in a heterohexameric (αβ) cage-like protein complex under ATP consumption. Here, we suggest ...The molybdenum storage protein (MoSto) deposits large amounts of molybdenum as polyoxomolybdate clusters in a heterohexameric (αβ) cage-like protein complex under ATP consumption. Here, we suggest a unique mechanism for the ATP-powered molybdate pumping process based on X-ray crystallography, cryoelectron microscopy, hydrogen-deuterium exchange mass spectrometry, and mutational studies of MoSto from . First, we show that molybdate, ATP, and Mg consecutively bind into the open ATP-binding groove of the β-subunit, which thereafter becomes tightly locked by fixing the previously disordered N-terminal arm of the α-subunit over the β-ATP. Next, we propose a nucleophilic attack of molybdate onto the γ-phosphate of β-ATP, analogous to the similar reaction of the structurally related UMP kinase. The formed instable phosphoric-molybdic anhydride becomes immediately hydrolyzed and, according to the current data, the released and accelerated molybdate is pressed through the cage wall, presumably by turning aside the Metβ149 side chain. A structural comparison between MoSto and UMP kinase provides valuable insight into how an enzyme is converted into a molecular machine during evolution. The postulated direct conversion of chemical energy into kinetic energy via an activating molybdate kinase and an exothermic pyrophosphatase reaction to overcome a proteinous barrier represents a novelty in ATP-fueled biochemistry, because normally, ATP hydrolysis initiates large-scale conformational changes to drive a distant process. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4907.map.gz emd_4907.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4907-v30.xml emd-4907-v30.xml emd-4907.xml emd-4907.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4907_fsc.xml emd_4907_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4907.png emd_4907.png | 215.5 KB | ||
| Masks |  emd_4907_msk_1.map emd_4907_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4907.cif.gz emd-4907.cif.gz | 6.7 KB | ||
| Others |  emd_4907_half_map_1.map.gz emd_4907_half_map_1.map.gz emd_4907_half_map_2.map.gz emd_4907_half_map_2.map.gz | 23.3 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4907 http://ftp.pdbj.org/pub/emdb/structures/EMD-4907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4907 | HTTPS FTP |
-Related structure data
| Related structure data |  6rkdMC  6risC  6rj4C  6rkeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4907.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4907.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | molybdenum storage protein under turnover conditions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4907_msk_1.map emd_4907_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: MoSto half map 1
| File | emd_4907_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MoSto half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: MoSto half map 2
| File | emd_4907_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MoSto half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer of A3B3 heterohexamer of molybdenum storage protein
| Entire | Name: Dimer of A3B3 heterohexamer of molybdenum storage protein |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of A3B3 heterohexamer of molybdenum storage protein
| Supramolecule | Name: Dimer of A3B3 heterohexamer of molybdenum storage protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) |
| Molecular weight | Theoretical: 380 KDa |
-Macromolecule #1: Molybdenum storage protein subunit alpha
| Macromolecule | Name: Molybdenum storage protein subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (strain DJ / ATCC BAA-1303) (bacteria) Azotobacter vinelandii (strain DJ / ATCC BAA-1303) (bacteria) |
| Molecular weight | Theoretical: 29.376773 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDTTNSIKH VISPLARQTL QDRDLTRPVA GKRPIRLLPW LQVVKIGGRV MDRGADAILP LVEELRKLLP EHRLLILTGA GVRARHVFS VGLDLGLPVG SLAPLAASEA GQNGHILAAM LASEGVSYVE HPTVADQLAI HLSATRAVVG SAFPPYHHHE F PGSRIPPH ...String: MTDTTNSIKH VISPLARQTL QDRDLTRPVA GKRPIRLLPW LQVVKIGGRV MDRGADAILP LVEELRKLLP EHRLLILTGA GVRARHVFS VGLDLGLPVG SLAPLAASEA GQNGHILAAM LASEGVSYVE HPTVADQLAI HLSATRAVVG SAFPPYHHHE F PGSRIPPH RADTGAFLLA DAFGAAGLTI VENVDGIYTA DPNGPDRGQA RFLPETSATD LAKSEGPLPV DRALLDVMAT AR HIERVQV VNGLVPGRLT AALRGEHVGT LIRTGVRPA UniProtKB: Molybdenum storage protein subunit alpha |
-Macromolecule #2: Molybdenum storage protein subunit beta
| Macromolecule | Name: Molybdenum storage protein subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (strain DJ / ATCC BAA-1303) (bacteria) Azotobacter vinelandii (strain DJ / ATCC BAA-1303) (bacteria) |
| Molecular weight | Theoretical: 28.378775 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANSTAELEE LLMQRSLTDP QLQAAAAAAA DFRILPDATV IKIGGQSVID RGRAAVYPLV DEIVAARKNH KLLIGTGAGT RARHLYSIA AGLGLPAGVL AQLGSSVADQ NAAMLGQLLA KHGIPVVGGA GLSAVPLSLA EVNAVVFSGM PPYKLWMRPA A EGVIPPYR ...String: MANSTAELEE LLMQRSLTDP QLQAAAAAAA DFRILPDATV IKIGGQSVID RGRAAVYPLV DEIVAARKNH KLLIGTGAGT RARHLYSIA AGLGLPAGVL AQLGSSVADQ NAAMLGQLLA KHGIPVVGGA GLSAVPLSLA EVNAVVFSGM PPYKLWMRPA A EGVIPPYR TDAGCFLLAE QFGCKQMIFV KDEDGLYTAN PKTSKDATFI PRISVDEMKA KGLHDSILEF PVLDLLQSAQ HV REVQVVN GLVPGNLTRA LAGEHVGTII TAS UniProtKB: Molybdenum storage protein subunit beta |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 12 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: bis(mu4-oxo)-tetrakis(mu3-oxo)-hexakis(mu2-oxo)-hexadecaoxo-octam...
| Macromolecule | Name: bis(mu4-oxo)-tetrakis(mu3-oxo)-hexakis(mu2-oxo)-hexadecaoxo-octamolybdenum (VI) type: ligand / ID: 5 / Number of copies: 12 / Formula: 8M0 |
|---|---|
| Molecular weight | Theoretical: 1.215503 KDa |
| Chemical component information | 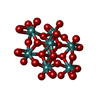 ChemComp-8M0: |
-Macromolecule #6: oxidanyl-[[2,2,4,4,4-pentakis($l^{1}-oxidanyl)-1-(oxidanylmolybde...
| Macromolecule | Name: oxidanyl-[[2,2,4,4,4-pentakis($l^{1}-oxidanyl)-1-(oxidanylmolybdenio)-1$l^{3},3-dioxa-2$l^{5},4$l^{5}-dimolybdacyclobut-2-yl]oxy]molybdenum type: ligand / ID: 6 / Number of copies: 12 / Formula: J8E |
|---|---|
| Molecular weight | Theoretical: 545.77 Da |
| Chemical component information | 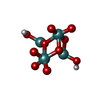 ChemComp-J8E: |
-Macromolecule #7: MOLYBDATE ION
| Macromolecule | Name: MOLYBDATE ION / type: ligand / ID: 7 / Number of copies: 42 / Formula: MOO |
|---|---|
| Molecular weight | Theoretical: 159.938 Da |
| Chemical component information | 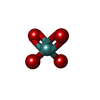 ChemComp-MOO: |
-Macromolecule #8: MO(VI)(=O)(OH)2 CLUSTER
| Macromolecule | Name: MO(VI)(=O)(OH)2 CLUSTER / type: ligand / ID: 8 / Number of copies: 6 / Formula: OMO |
|---|---|
| Molecular weight | Theoretical: 145.954 Da |
| Chemical component information |  ChemComp-OMO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 6.5 / Component - Concentration: 50.0 mM / Component - Name: MOPS/NaOH Details: 1 mM molybdate and 1 mM mg-ATP were added before vitrification. |
| Grid | Model: C-flat / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 1238 / Average exposure time: 8.0 sec. / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.9 µm / Calibrated magnification: 45045 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: JEOL / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Phenix_real_space_refine | ||||||
| Refinement | Space: REAL / Protocol: OTHER | ||||||
| Output model |  PDB-6rkd: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)