+Search query
-Structure paper
| Title | Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 116, Issue 52, Page 26497-26504, Year 2019 |
| Publish date | Dec 26, 2019 |
 Authors Authors | Steffen Brünle / Martin L Eisinger / Juliane Poppe / Deryck J Mills / Julian D Langer / Janet Vonck / Ulrich Ermler /  |
| PubMed Abstract | The molybdenum storage protein (MoSto) deposits large amounts of molybdenum as polyoxomolybdate clusters in a heterohexameric (αβ) cage-like protein complex under ATP consumption. Here, we suggest ...The molybdenum storage protein (MoSto) deposits large amounts of molybdenum as polyoxomolybdate clusters in a heterohexameric (αβ) cage-like protein complex under ATP consumption. Here, we suggest a unique mechanism for the ATP-powered molybdate pumping process based on X-ray crystallography, cryoelectron microscopy, hydrogen-deuterium exchange mass spectrometry, and mutational studies of MoSto from . First, we show that molybdate, ATP, and Mg consecutively bind into the open ATP-binding groove of the β-subunit, which thereafter becomes tightly locked by fixing the previously disordered N-terminal arm of the α-subunit over the β-ATP. Next, we propose a nucleophilic attack of molybdate onto the γ-phosphate of β-ATP, analogous to the similar reaction of the structurally related UMP kinase. The formed instable phosphoric-molybdic anhydride becomes immediately hydrolyzed and, according to the current data, the released and accelerated molybdate is pressed through the cage wall, presumably by turning aside the Metβ149 side chain. A structural comparison between MoSto and UMP kinase provides valuable insight into how an enzyme is converted into a molecular machine during evolution. The postulated direct conversion of chemical energy into kinetic energy via an activating molybdate kinase and an exothermic pyrophosphatase reaction to overcome a proteinous barrier represents a novelty in ATP-fueled biochemistry, because normally, ATP hydrolysis initiates large-scale conformational changes to drive a distant process. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:31811022 / PubMed:31811022 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.7 - 3.2 Å |
| Structure data | EMDB-4907, PDB-6rkd:  PDB-6ris:  PDB-6rj4:  PDB-6rke: |
| Chemicals |  ChemComp-ATP:  ChemComp-PO4:  ChemComp-MG:  ChemComp-CL:  ChemComp-HOH:  ChemComp-ADP:  ChemComp-NA: 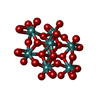 ChemComp-8M0: 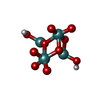 ChemComp-J8E: 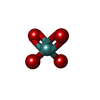 ChemComp-MOO:  ChemComp-OMO: 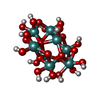 ChemComp-LJB:  ChemComp-M10:  ChemComp-LHW: |
| Source |
|
 Keywords Keywords | METAL BINDING PROTEIN / amino acid kinase family / Mo storage / polyoxomolybdate cluster / ATP hydrolysis / polyoxomolybdate clusters / amino acid kinase / molybdenum storage protein / ATPase / ATP |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 azotobacter vinelandii dj (bacteria)
azotobacter vinelandii dj (bacteria)