[English] 日本語
 Yorodumi
Yorodumi- EMDB-4905: 3D structure of horse spleen apoferritin determined using multifu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4905 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3D structure of horse spleen apoferritin determined using multifunctional graphene supports for electron cryomicroscopy | |||||||||
 Map data Map data | Post-processed, masked and filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | METAL TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationferritin complex / autolysosome / ferric iron binding / autophagosome / iron ion transport / ferrous iron binding / cytoplasmic vesicle / intracellular iron ion homeostasis / iron ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.1 Å | |||||||||
 Authors Authors | Naydenova K / Peet MJ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Multifunctional graphene supports for electron cryomicroscopy. Authors: Katerina Naydenova / Mathew J Peet / Christopher J Russo /  Abstract: With recent technological advances, the atomic resolution structure of any purified biomolecular complex can, in principle, be determined by single-particle electron cryomicroscopy (cryoEM). In ...With recent technological advances, the atomic resolution structure of any purified biomolecular complex can, in principle, be determined by single-particle electron cryomicroscopy (cryoEM). In practice, the primary barrier to structure determination is the preparation of a frozen specimen suitable for high-resolution imaging. To address this, we present a multifunctional specimen support for cryoEM, comprising large-crystal monolayer graphene suspended across the surface of an ultrastable gold specimen support. Using a low-energy plasma surface modification system, we tune the surface of this support to the specimen by patterning a range of covalent functionalizations across the graphene layer on a single grid. This support design reduces specimen movement during imaging, improves image quality, and allows high-resolution structure determination with a minimum of material and data. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4905.map.gz emd_4905.map.gz | 22.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4905-v30.xml emd-4905-v30.xml emd-4905.xml emd-4905.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4905_fsc.xml emd_4905_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4905.png emd_4905.png | 129.8 KB | ||
| Masks |  emd_4905_msk_1.map emd_4905_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4905.cif.gz emd-4905.cif.gz | 5.3 KB | ||
| Others |  emd_4905_half_map_1.map.gz emd_4905_half_map_1.map.gz emd_4905_half_map_2.map.gz emd_4905_half_map_2.map.gz | 113.5 MB 113.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4905 http://ftp.pdbj.org/pub/emdb/structures/EMD-4905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4905 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4905 | HTTPS FTP |
-Related structure data
| Related structure data |  6rjhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10272 (Title: Movies of horse spleen apoferritin on multifunctional ultrastable graphene supports for electron cryomicroscopy EMPIAR-10272 (Title: Movies of horse spleen apoferritin on multifunctional ultrastable graphene supports for electron cryomicroscopyData size: 570.0 Data #1: Movies of apoferritin on functionalized ultrastable graphene support [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4905.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4905.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed, masked and filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.6495 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4905_msk_1.map emd_4905_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
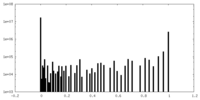
| Density Histograms |
-Half map: Half map 1
| File | emd_4905_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
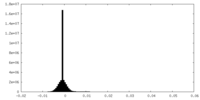
| Density Histograms |
-Half map: Half map 2
| File | emd_4905_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
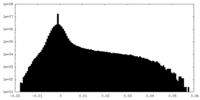
| Density Histograms |
- Sample components
Sample components
-Entire : Apoferritin
| Entire | Name: Apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: Apoferritin
| Supramolecule | Name: Apoferritin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Ferritin light chain
| Macromolecule | Name: Ferritin light chain / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.532113 KDa |
| Sequence | String: SQIRQNYSTE VEAAVNRLVN LYLRASYTYL SLGFYFDRDD VALEGVCHFF RELAEEKREG AERLLKMQNQ RGGRALFQDL QKPSQDEWG TTLDAMKAAI VLEKSLNQAL LDLHALGSAQ ADPHLCDFLE SHFLDEEVKL IKKMGDHLTN IQRLVGSQAG L GEYLFERL TLK UniProtKB: Ferritin light chain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Homemade |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 37.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)