[English] 日本語
 Yorodumi
Yorodumi- EMDB-47952: Cryo-EM structure of the adenosine A2A receptor intermediate boun... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the adenosine A2A receptor intermediate bound to a miniGs heterotrimer | |||||||||
 Map data Map data | Raw Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Heterotrimer / Receptor / cryo-EM / SIGNALING PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of norepinephrine secretion / positive regulation of acetylcholine secretion, neurotransmission / negative regulation of alpha-beta T cell activation / positive regulation of circadian sleep/wake cycle, sleep / Adenosine P1 receptors / G protein-coupled adenosine receptor activity / response to purine-containing compound / G protein-coupled adenosine receptor signaling pathway / NGF-independant TRKA activation / Surfactant metabolism ...regulation of norepinephrine secretion / positive regulation of acetylcholine secretion, neurotransmission / negative regulation of alpha-beta T cell activation / positive regulation of circadian sleep/wake cycle, sleep / Adenosine P1 receptors / G protein-coupled adenosine receptor activity / response to purine-containing compound / G protein-coupled adenosine receptor signaling pathway / NGF-independant TRKA activation / Surfactant metabolism / sensory perception / positive regulation of urine volume / synaptic transmission, dopaminergic / type 5 metabotropic glutamate receptor binding / negative regulation of vascular permeability / synaptic transmission, cholinergic / positive regulation of glutamate secretion / intermediate filament / presynaptic active zone / blood circulation / response to caffeine / eating behavior / inhibitory postsynaptic potential / alpha-actinin binding / regulation of calcium ion transport / asymmetric synapse / PKA activation in glucagon signalling / axolemma / developmental growth / membrane depolarization / hair follicle placode formation / phagocytosis / D1 dopamine receptor binding / cellular defense response / intracellular transport / prepulse inhibition / positive regulation of synaptic transmission, glutamatergic / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / neuron projection morphogenesis / adenylate cyclase-activating adrenergic receptor signaling pathway / astrocyte activation / regulation of insulin secretion / presynaptic modulation of chemical synaptic transmission / cellular response to glucagon stimulus / response to amphetamine / central nervous system development / adenylate cyclase activator activity / trans-Golgi network membrane / positive regulation of long-term synaptic potentiation / positive regulation of synaptic transmission, GABAergic / positive regulation of protein secretion / positive regulation of apoptotic signaling pathway / regulation of mitochondrial membrane potential / excitatory postsynaptic potential / synaptic transmission, glutamatergic / apoptotic signaling pathway / negative regulation of inflammatory response to antigenic stimulus / locomotory behavior / bone development / negative regulation of inflammatory response / platelet aggregation / vasodilation / cognition / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / blood coagulation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / cell-cell signaling Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.16 Å | |||||||||
 Authors Authors | Bi M / Wang X / Ye L / Cheng Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structure and function of a near fully-activated intermediate GPCR-Gαβγ complex. Authors: Maxine Bi / Xudong Wang / Jinan Wang / Jun Xu / Wenkai Sun / Victor Ayo Adediwura / Yinglong Miao / Yifan Cheng / Libin Ye /  Abstract: Unraveling the signaling roles of intermediate complexes is pivotal for G protein-coupled receptor (GPCR) drug development. Despite hundreds of GPCR-Gαβγ structures, these snapshots primarily ...Unraveling the signaling roles of intermediate complexes is pivotal for G protein-coupled receptor (GPCR) drug development. Despite hundreds of GPCR-Gαβγ structures, these snapshots primarily capture the fully activated complex. Consequently, the functions of intermediate GPCR-G protein complexes remain elusive. Guided by a conformational landscape visualized via F quantitative NMR and molecular dynamics (MD) simulations, we determined the structure of an intermediate GPCR-mini-Gαβγ complex at 2.6 Å using cryo-EM, by blocking its transition to the fully activated complex. Furthermore, we present direct evidence that the complex at this intermediate state initiates a rate-limited nucleotide exchange before transitioning to the fully activated complex. In this state, BODIPY-GDP/GTP based nucleotide exchange assays further indicated the α-helical domain of the Gα is partially open, allowing it to grasp a nucleotide at a non-canonical binding site, distinct from the canonical nucleotide-binding site. These advances bridge a significant gap in our understanding of the complexity of GPCR signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47952.map.gz emd_47952.map.gz | 109.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47952-v30.xml emd-47952-v30.xml emd-47952.xml emd-47952.xml | 27.1 KB 27.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_47952.png emd_47952.png | 40 KB | ||
| Masks |  emd_47952_msk_1.map emd_47952_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-47952.cif.gz emd-47952.cif.gz | 7.3 KB | ||
| Others |  emd_47952_additional_1.map.gz emd_47952_additional_1.map.gz emd_47952_half_map_1.map.gz emd_47952_half_map_1.map.gz emd_47952_half_map_2.map.gz emd_47952_half_map_2.map.gz | 109.8 MB 92.9 MB 94.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47952 http://ftp.pdbj.org/pub/emdb/structures/EMD-47952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47952 | HTTPS FTP |
-Related structure data
| Related structure data |  9ee9MC  9ee8C  9eeaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47952.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47952.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
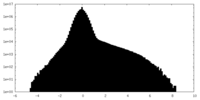
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_47952_msk_1.map emd_47952_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened Map
| File | emd_47952_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map1
| File | emd_47952_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map2
| File | emd_47952_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the GPCR bound to a miniGs heterotrimer
| Entire | Name: Cryo-EM structure of the GPCR bound to a miniGs heterotrimer |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the GPCR bound to a miniGs heterotrimer
| Supramolecule | Name: Cryo-EM structure of the GPCR bound to a miniGs heterotrimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 144 KDa |
-Macromolecule #1: Adenosine receptor A2a
| Macromolecule | Name: Adenosine receptor A2a / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.517078 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: DYKDDDDKSN NNNNNNNNLG ENLYFQGAPI MGSSVYITVE LAIAVLAILG NVLVCWAVWL NSNLQNVTNY FVVSLAAADI AVGVLAIPF AITISTGFCA ACHGCLFIAC FVLVLTQSSI FSLLAIAIDR YIAIRIPLRY NGLVTGTRAK GIIAICWVLS F AIGLTPML ...String: DYKDDDDKSN NNNNNNNNLG ENLYFQGAPI MGSSVYITVE LAIAVLAILG NVLVCWAVWL NSNLQNVTNY FVVSLAAADI AVGVLAIPF AITISTGFCA ACHGCLFIAC FVLVLTQSSI FSLLAIAIDR YIAIRIPLRY NGLVTGTRAK GIIAICWVLS F AIGLTPML GWNNCGQPKE GKNHSQGCGE GQVACLFEDV VPMNYMVYFN FFACVLVPLL LMLGVYLRIF LAARRQLKQM ES QPLPGER ARSTLQKECH AAKSLAIIVG LFALCWLPLH IINCFTFFCP DCSHAPLWLM YLAIVLSHTN SVVNPFIYAY AIR EFRQTF RKIIRSHVLR QQEPFKAHHH HHHHHHH UniProtKB: Adenosine receptor A2a |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.728152 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSSGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: GPGSSGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.776713 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHEN LYFQGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGADN SGKSTIVKQM RILHGGSGGS GGTSGIFET KFQVDKVNFH MFDVGGQRDE RRKWIQCFND VTAIIFVVDS SDYNRLQEAL NLFKSIWNNR WLRTISVILF L NKQDLLAE ...String: MGHHHHHHEN LYFQGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGADN SGKSTIVKQM RILHGGSGGS GGTSGIFET KFQVDKVNFH MFDVGGQRDE RRKWIQCFND VTAIIFVVDS SDYNRLQEAL NLFKSIWNNR WLRTISVILF L NKQDLLAE KVLAGKSKIE DYFPEFARYT TPEDATPEPG EDPRVTRAKY FIRDEFLRIS TASGDGRHYC YPHFTCAVDT EN ARRIFND CRDIIQRMHL RQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #5: nanobody Nb35
| Macromolecule | Name: nanobody Nb35 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.926076 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAQVQLQESG GGLVQPGGSL RLSCAASGFT FSNYKMNWVR QAPGKGLEWV SDISQSGASI SYTGSVKGR FTISRDNAKN TLYLQMNSLK PEDTAVYYCA RCPAPFTRDC FDVTSTTYAY RGQGTQVTVS SHHHHHH |
-Macromolecule #6: ADENOSINE
| Macromolecule | Name: ADENOSINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: ADN |
|---|---|
| Molecular weight | Theoretical: 267.241 Da |
| Chemical component information |  ChemComp-ADN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 47.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)