[English] 日本語
 Yorodumi
Yorodumi- EMDB-4749: Multidrug resistance transporter BmrA mutant E504A bound with ATP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4749 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Multidrug resistance transporter BmrA mutant E504A bound with ATP and Mg solved by Cryo-EM | ||||||||||||
 Map data Map data | Main map used for model building. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | membrane protein / ABC transporter / multidrug resistance / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationATPase-coupled lipid transmembrane transporter activity / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / ABC-type transporter activity / transmembrane transport / response to antibiotic / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
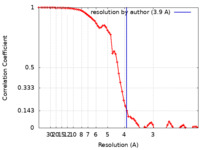
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Wiseman B / Chaptal V | ||||||||||||
| Funding support |  France, France,  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter. Authors: Vincent Chaptal / Veronica Zampieri / Benjamin Wiseman / Cédric Orelle / Juliette Martin / Kim-Anh Nguyen / Alexia Gobet / Margot Di Cesare / Sandrine Magnard / Waqas Javed / Jad Eid / ...Authors: Vincent Chaptal / Veronica Zampieri / Benjamin Wiseman / Cédric Orelle / Juliette Martin / Kim-Anh Nguyen / Alexia Gobet / Margot Di Cesare / Sandrine Magnard / Waqas Javed / Jad Eid / Arnaud Kilburg / Marine Peuchmaur / Julien Marcoux / Luca Monticelli / Martin Hogbom / Guy Schoehn / Jean-Michel Jault / Ahcène Boumendjel / Pierre Falson /   Abstract: Multidrug ABC transporters translocate drugs across membranes by a mechanism for which the molecular features of drug release are so far unknown. Here, we resolved three ATP-Mg-bound outward-facing ...Multidrug ABC transporters translocate drugs across membranes by a mechanism for which the molecular features of drug release are so far unknown. Here, we resolved three ATP-Mg-bound outward-facing conformations of the (homodimeric) BmrA by x-ray crystallography and single-particle cryo-electron microscopy (EM) in detergent solution, one of them with rhodamine 6G (R6G), a substrate exported by BmrA when overexpressed in . Two R6G molecules bind to the drug-binding cavity at the level of the outer leaflet, between transmembrane (TM) helices 1-2 of one monomer and TM5'-6' of the other. They induce a rearrangement of TM1-2, highlighting a local flexibility that we confirmed by hydrogen/deuterium exchange and molecular dynamics simulations. In the absence of R6G, simulations show a fast postrelease occlusion of the cavity driven by hydrophobicity, while when present, R6G can move within the cavity, maintaining it open. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4749.map.gz emd_4749.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4749-v30.xml emd-4749-v30.xml emd-4749.xml emd-4749.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4749_fsc.xml emd_4749_fsc.xml | 7.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_4749.png emd_4749.png | 54 KB | ||
| Filedesc metadata |  emd-4749.cif.gz emd-4749.cif.gz | 6.6 KB | ||
| Others |  emd_4749_additional.map.gz emd_4749_additional.map.gz emd_4749_additional_1.map.gz emd_4749_additional_1.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4749 http://ftp.pdbj.org/pub/emdb/structures/EMD-4749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4749 | HTTPS FTP |
-Related structure data
| Related structure data |  6r81MC  6r72C  7bg4C  7ow8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4749.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4749.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map used for model building. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened map used to aid and improve model building in many areas.
| File | emd_4749_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map used to aid and improve model building in many areas. | ||||||||||||
| Projections & Slices |
| ||||||||||||
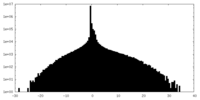
| Density Histograms |
-Additional map: Sharpened map used to aid and improve model building in many areas.
| File | emd_4749_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map used to aid and improve model building in many areas. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Multidrug resistance transporter from Bacillus subtilis bound wit...
| Entire | Name: Multidrug resistance transporter from Bacillus subtilis bound with ATP and Mg. |
|---|---|
| Components |
|
-Supramolecule #1: Multidrug resistance transporter from Bacillus subtilis bound wit...
| Supramolecule | Name: Multidrug resistance transporter from Bacillus subtilis bound with ATP and Mg. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: mutant E504A |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Lipid A export ATP-binding/permease protein MsbA
| Macromolecule | Name: Lipid A export ATP-binding/permease protein MsbA / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.747141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSSHHHHHH MPTKKQKSKS KLKPFFALVR RTNPSYGKLA FALALSVVTT LVSLLIPLLT KQLVDGFSMS NLSGTQIGLI ALVFFVQAG LSAYATYALN YNGQKIISGL RELLWKKLIK LPVSYFDTNA SGETVSRVTN DTMVVKELIT THISGFITGI I SVIGSLTI ...String: MSSSHHHHHH MPTKKQKSKS KLKPFFALVR RTNPSYGKLA FALALSVVTT LVSLLIPLLT KQLVDGFSMS NLSGTQIGLI ALVFFVQAG LSAYATYALN YNGQKIISGL RELLWKKLIK LPVSYFDTNA SGETVSRVTN DTMVVKELIT THISGFITGI I SVIGSLTI LFIMNWKLTL LVLVVVPLAA LILVPIGRKM FSISRETQDE TARFTGLLNQ ILPEIRLVKA SNAEDVEYGR GK MGISSLF KLGVREAKVQ SLVGPLISLV LMAALVAVIG YGGMQVSSGE LTAGALVAFI LYLFQIIMPM GQITTFFTQL QKS IGATER MIEILAEEEE DTVTGKQIEN AHLPIQLDRV SFGYKPDQLI LKEVSAVIEA GKVTAIVGPS GGGKTTLFKL LERF YSPTA GTIRLGDEPV DTYSLESWRE HIGYVSQESP LMSGTIRENI CYGLERDVTD AEIEKAAEMA YALNFIKELP NQFDT EVGE RGIMLSGGQR QRIAIARALL RNPSILMLDA ATSSLDSQSE KSVQQALEVL MEGRTTIVIA HRLSTVVDAD QLLFVE KGE ITGRGTHHEL MASHGLYRDF AEQQLKMNAD LENKAG UniProtKB: Lipid A export ATP-binding/permease protein MsbA |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3477 / Average exposure time: 5.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 187 |
|---|---|
| Output model |  PDB-6r81: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)