[English] 日本語
 Yorodumi
Yorodumi- EMDB-0606: Complex of human cystic fibrosis transmembrane conductance regula... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0606 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of human cystic fibrosis transmembrane conductance regulator (CFTR) and GLPG1837 | |||||||||
 Map data Map data | B-factor sharpened map at -50A2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / anion channel / cystic fibrosis / membrane protein / GLPG1837 / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of voltage-gated chloride channel activity / : / Sec61 translocon complex binding / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / positive regulation of enamel mineralization / transepithelial water transport / RHO GTPases regulate CFTR trafficking / amelogenesis / intracellular pH elevation ...positive regulation of voltage-gated chloride channel activity / : / Sec61 translocon complex binding / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / positive regulation of enamel mineralization / transepithelial water transport / RHO GTPases regulate CFTR trafficking / amelogenesis / intracellular pH elevation / chloride channel inhibitor activity / : / multicellular organismal-level water homeostasis / Golgi-associated vesicle membrane / chloride channel regulator activity / cholesterol transport / bicarbonate transport / bicarbonate transmembrane transporter activity / membrane hyperpolarization / vesicle docking involved in exocytosis / chloride transmembrane transporter activity / cholesterol biosynthetic process / sperm capacitation / RHOQ GTPase cycle / chloride channel activity / positive regulation of exocytosis / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / chloride channel complex / 14-3-3 protein binding / cellular response to forskolin / chloride transmembrane transport / cellular response to cAMP / response to endoplasmic reticulum stress / PDZ domain binding / establishment of localization in cell / clathrin-coated endocytic vesicle membrane / Defective CFTR causes cystic fibrosis / Late endosomal microautophagy / recycling endosome / ABC-family proteins mediated transport / transmembrane transport / recycling endosome membrane / Chaperone Mediated Autophagy / Aggrephagy / Cargo recognition for clathrin-mediated endocytosis / protein-folding chaperone binding / Clathrin-mediated endocytosis / early endosome membrane / early endosome / apical plasma membrane / endosome membrane / Ub-specific processing proteases / lysosomal membrane / endoplasmic reticulum membrane / enzyme binding / cell surface / ATP hydrolysis activity / protein-containing complex / ATP binding / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Zhang Z / Liu F | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Structural identification of a hotspot on CFTR for potentiation. Authors: Fangyu Liu / Zhe Zhang / Anat Levit / Jesper Levring / Kouki K Touhara / Brian K Shoichet / Jue Chen /  Abstract: Cystic fibrosis is a fatal disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR). Two main categories of drugs are being developed: correctors that improve ...Cystic fibrosis is a fatal disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR). Two main categories of drugs are being developed: correctors that improve folding of CFTR and potentiators that recover the function of CFTR. Here, we report two cryo-electron microscopy structures of human CFTR in complex with potentiators: one with the U.S. Food and Drug Administration (FDA)-approved drug ivacaftor at 3.3-angstrom resolution and the other with an investigational drug, GLPG1837, at 3.2-angstrom resolution. These two drugs, although chemically dissimilar, bind to the same site within the transmembrane region. Mutagenesis suggests that in both cases, hydrogen bonds provided by the protein are important for drug recognition. The molecular details of how ivacaftor and GLPG1837 interact with CFTR may facilitate structure-based optimization of therapeutic compounds. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0606.map.gz emd_0606.map.gz | 95.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0606-v30.xml emd-0606-v30.xml emd-0606.xml emd-0606.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0606.png emd_0606.png | 51.5 KB | ||
| Filedesc metadata |  emd-0606.cif.gz emd-0606.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0606 http://ftp.pdbj.org/pub/emdb/structures/EMD-0606 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0606 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0606 | HTTPS FTP |
-Related structure data
| Related structure data |  6o1vMC  0611C  6o2pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10281 (Title: Cryo electron micrographs of human cystic fibrosis transmembrane conductance regulator (CFTR) in complex with GLPG EMPIAR-10281 (Title: Cryo electron micrographs of human cystic fibrosis transmembrane conductance regulator (CFTR) in complex with GLPGData size: 1.4 TB Data #1: Unaligned multi-frame micrographs of human CFTR in complex with GLPG1837 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0606.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0606.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map at -50A2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of human cystic fibrosis transmembrane conductance regula...
| Entire | Name: Complex of human cystic fibrosis transmembrane conductance regulator (CFTR) and GLPG1837 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of human cystic fibrosis transmembrane conductance regula...
| Supramolecule | Name: Complex of human cystic fibrosis transmembrane conductance regulator (CFTR) and GLPG1837 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 168 KDa |
-Macromolecule #1: Cystic fibrosis transmembrane conductance regulator
| Macromolecule | Name: Cystic fibrosis transmembrane conductance regulator / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ec: 3.6.3.49 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 169.352594 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQRSPLEKAS VVSKLFFSWT RPILRKGYRQ RLELSDIYQI PSVDSADNLS EKLEREWDRE LASKKNPKLI NALRRCFFWR FMFYGIFLY LGEVTKAVQP LLLGRIIASY DPDNKEERSI AIYLGIGLCL LFIVRTLLLH PAIFGLHHIG MQMRIAMFSL I YKKTLKLS ...String: MQRSPLEKAS VVSKLFFSWT RPILRKGYRQ RLELSDIYQI PSVDSADNLS EKLEREWDRE LASKKNPKLI NALRRCFFWR FMFYGIFLY LGEVTKAVQP LLLGRIIASY DPDNKEERSI AIYLGIGLCL LFIVRTLLLH PAIFGLHHIG MQMRIAMFSL I YKKTLKLS SRVLDKISIG QLVSLLSNNL NKFDEGLALA HFVWIAPLQV ALLMGLIWEL LQASAFCGLG FLIVLALFQA GL GRMMMKY RDQRAGKISE RLVITSEMIE NIQSVKAYCW EEAMEKMIEN LRQTELKLTR KAAYVRYFNS SAFFFSGFFV VFL SVLPYA LIKGIILRKI FTTISFCIVL RMAVTRQFPW AVQTWYDSLG AINKIQDFLQ KQEYKTLEYN LTTTEVVMEN VTAF WEEGF GELFEKAKQN NNNRKTSNGD DSLFFSNFSL LGTPVLKDIN FKIERGQLLA VAGSTGAGKT SLLMVIMGEL EPSEG KIKH SGRISFCSQF SWIMPGTIKE NIIFGVSYDE YRYRSVIKAC QLEEDISKFA EKDNIVLGEG GITLSGGQRA RISLAR AVY KDADLYLLDS PFGYLDVLTE KEIFESCVCK LMANKTRILV TSKMEHLKKA DKILILHEGS SYFYGTFSEL QNLQPDF SS KLMGCDSFDQ FSAERRNSIL TETLHRFSLE GDAPVSWTET KKQSFKQTGE FGEKRKNSIL NPINSIRKFS IVQKTPLQ M NGIEEDSDEP LERRLSLVPD SEQGEAILPR ISVISTGPTL QARRRQSVLN LMTHSVNQGQ NIHRKTTAST RKVSLAPQA NLTELDIYSR RLSQETGLEI SEEINEEDLK ECFFDDMESI PAVTTWNTYL RYITVHKSLI FVLIWCLVIF LAEVAASLVV LWLLGNTPL QDKGNSTHSR NNSYAVIITS TSSYYVFYIY VGVADTLLAM GFFRGLPLVH TLITVSKILH HKMLHSVLQA P MSTLNTLK AGGILNRFSK DIAILDDLLP LTIFDFIQLL LIVIGAIAVV AVLQPYIFVA TVPVIVAFIM LRAYFLQTSQ QL KQLESEG RSPIFTHLVT SLKGLWTLRA FGRQPYFETL FHKALNLHTA NWFLYLSTLR WFQMRIEMIF VIFFIAVTFI SIL TTGEGE GRVGIILTLA MNIMSTLQWA VNSSIDVDSL MRSVSRVFKF IDMPTEGKPT KSTKPYKNGQ LSKVMIIENS HVKK DDIWP SGGQMTVKDL TAKYTEGGNA ILENISFSIS PGQRVGLLGR TGSGKSTLLS AFLRLLNTEG EIQIDGVSWD SITLQ QWRK AFGVIPQKVF IFSGTFRKNL DPYEQWSDQE IWKVADEVGL RSVIEQFPGK LDFVLVDGGC VLSHGHKQLM CLARSV LSK AKILLLDQPS AHLDPVTYQI IRRTLKQAFA DCTVILCEHR IEAMLECQQF LVIEENKVRQ YDSIQKLLNE RSLFRQA IS PSDRVKLFPH RNSSKCKSKP QIAALKEETE EEVQDTRLSN SLEVLFQ UniProtKB: Cystic fibrosis transmembrane conductance regulator |
-Macromolecule #2: Unknown Peptide
| Macromolecule | Name: Unknown Peptide / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.464797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #3: N-(3-carbamoyl-5,5,7,7-tetramethyl-4,7-dihydro-5H-thieno[2,3-c]py...
| Macromolecule | Name: N-(3-carbamoyl-5,5,7,7-tetramethyl-4,7-dihydro-5H-thieno[2,3-c]pyran-2-yl)-1H-pyrazole-3-carboxamide type: ligand / ID: 3 / Number of copies: 1 / Formula: LJP |
|---|---|
| Molecular weight | Theoretical: 348.42 Da |
| Chemical component information | 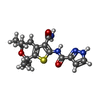 ChemComp-LJP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 6 / Number of copies: 5 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.51 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























