[English] 日本語
 Yorodumi
Yorodumi- EMDB-4666: Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrime... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4666 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | ||||||||||||
 Map data Map data | None | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Influenza A / RNA polymerase / Influenza polymerase / Influenza dimer / RDRP / RNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / virion component / endonuclease activity / Hydrolases; Acting on ester bonds / host cell cytoplasm ...cap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / virion component / endonuclease activity / Hydrolases; Acting on ester bonds / host cell cytoplasm / symbiont-mediated suppression of host gene expression / viral translational frameshifting / RNA-directed RNA polymerase / viral RNA genome replication / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / host cell nucleus / RNA binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Influenza A virus (A/nt/60/1968(H3N2)) / Influenza A virus (A/nt/60/1968(H3N2)) Influenza A virus (A/nt/60/1968(H3N2)) / Influenza A virus (A/nt/60/1968(H3N2)) | ||||||||||||
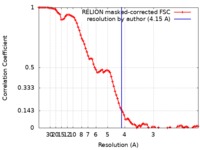
| Method | single particle reconstruction / cryo EM / Resolution: 4.15 Å | ||||||||||||
 Authors Authors | Carrique L / Keown JR | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structures of influenza A virus RNA polymerase offer insight into viral genome replication. Authors: Haitian Fan / Alexander P Walker / Loïc Carrique / Jeremy R Keown / Itziar Serna Martin / Dimple Karia / Jane Sharps / Narin Hengrung / Els Pardon / Jan Steyaert / Jonathan M Grimes / Ervin Fodor /    Abstract: Influenza A viruses are responsible for seasonal epidemics, and pandemics can arise from the transmission of novel zoonotic influenza A viruses to humans. Influenza A viruses contain a segmented ...Influenza A viruses are responsible for seasonal epidemics, and pandemics can arise from the transmission of novel zoonotic influenza A viruses to humans. Influenza A viruses contain a segmented negative-sense RNA genome, which is transcribed and replicated by the viral-RNA-dependent RNA polymerase (FluPol) composed of PB1, PB2 and PA subunits. Although the high-resolution crystal structure of FluPol of bat influenza A virus has previously been reported, there are no complete structures available for human and avian FluPol. Furthermore, the molecular mechanisms of genomic viral RNA (vRNA) replication-which proceeds through a complementary RNA (cRNA) replicative intermediate, and requires oligomerization of the polymerase-remain largely unknown. Here, using crystallography and cryo-electron microscopy, we determine the structures of FluPol from human influenza A/NT/60/1968 (H3N2) and avian influenza A/duck/Fujian/01/2002 (H5N1) viruses at a resolution of 3.0-4.3 Å, in the presence or absence of a cRNA or vRNA template. In solution, FluPol forms dimers of heterotrimers through the C-terminal domain of the PA subunit, the thumb subdomain of PB1 and the N1 subdomain of PB2. The cryo-electron microscopy structure of monomeric FluPol bound to the cRNA template reveals a binding site for the 3' cRNA at the dimer interface. We use a combination of cell-based and in vitro assays to show that the interface of the FluPol dimer is required for vRNA synthesis during replication of the viral genome. We also show that a nanobody (a single-domain antibody) that interferes with FluPol dimerization inhibits the synthesis of vRNA and, consequently, inhibits virus replication in infected cells. Our study provides high-resolution structures of medically relevant FluPol, as well as insights into the replication mechanisms of the viral RNA genome. In addition, our work identifies sites in FluPol that could be targeted in the development of antiviral drugs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4666.map.gz emd_4666.map.gz | 36.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4666-v30.xml emd-4666-v30.xml emd-4666.xml emd-4666.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4666_fsc.xml emd_4666_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_4666.png emd_4666.png | 67 KB | ||
| Filedesc metadata |  emd-4666.cif.gz emd-4666.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4666 http://ftp.pdbj.org/pub/emdb/structures/EMD-4666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4666 | HTTPS FTP |
-Related structure data
| Related structure data |  6qxeMC  4660C  4661C  4663C  4664C  4986C  6qnwC  6qpfC  6qpgC  6qwlC  6qx3C  6qx8C  6rr7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4666.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4666.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrime...
| Entire | Name: Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 |
|---|---|
| Components |
|
-Supramolecule #1: Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrime...
| Supramolecule | Name: Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Sample was treated with 0.001% glutaraldehyde for 20 min on ice prior quenching with 100 mM Tris-HCl pH 7.5 and gel filtration. |
|---|---|
| Molecular weight | Theoretical: 250 KDa |
-Supramolecule #2: Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrime...
| Supramolecule | Name: Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/nt/60/1968(H3N2)) Influenza A virus (A/nt/60/1968(H3N2)) |
-Supramolecule #3: Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrime...
| Supramolecule | Name: Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/nt/60/1968(H3N2)) Influenza A virus (A/nt/60/1968(H3N2)) |
-Macromolecule #1: Nb8205
| Macromolecule | Name: Nb8205 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Influenza A virus (A/nt/60/1968(H3N2)) |
| Molecular weight | Theoretical: 14.835375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG MVQPGGSLRL SCLASGFTFS NYAMTWVRQA PGKGPEWVSM VSNNGADTTY TDSVKGRFTI SRDNAKNTLY LRMNNVKPE DSAVYYCAKR RYGGIWTGQP TDYDYLGQGT VTVSSHHHHH HEPEA |
-Macromolecule #2: Polymerase acidic protein
| Macromolecule | Name: Polymerase acidic protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism: Influenza A virus (A/nt/60/1968(H3N2)) |
| Molecular weight | Theoretical: 83.100797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEDFVRQCFN PMIVELAEKA MKEYGEDLKI ETNKFAAICT HLEVCFMYSD FHFINEQGES IVVELDDPNA LLKHRFEIIE GRDRTMAWT VVNSICNTTG AEKPKFLPDL YDYKENRFIE IGVTRREVHI YYLEKANKIK SENTHIHIFS FTGEEMATKA D YTLDEESR ...String: MEDFVRQCFN PMIVELAEKA MKEYGEDLKI ETNKFAAICT HLEVCFMYSD FHFINEQGES IVVELDDPNA LLKHRFEIIE GRDRTMAWT VVNSICNTTG AEKPKFLPDL YDYKENRFIE IGVTRREVHI YYLEKANKIK SENTHIHIFS FTGEEMATKA D YTLDEESR ARIKTRLFTI RQEMANRGLW DSFRQSERGE ETIEERFEIT GTMRRLADQS LPPNFSCLEN FRAYVDGFEP NG YIEGKLS QMSKEVNAKI EPFLKTTPRP IRLPDGPPCF QRSKFLLMDA LKLSIEDPSH EGEGIPLYDA IKCMRTFFGW KEP YIVKPH EKGINPNYLL SWKQVLAELQ DIENEEKIPR TKNMKKTSQL KWALGENMAP EKVDFDNCRD VSDLKQYDSD EPEL RSLSS WIQNEFNKAC ELTDSTWIEL DEIGEDVAPI EYIASMRRNY FTAEVSHCRA TEYIMKGVYI NTALLNASCA AMDDF QLIP MISKCRTKEG RRKTNLYGFI IKGRSHLRND TDVVNFVSME FSLTDPRLEP HKWEKYCVLE IGDMLLRSAI GQMSRP MFL YVRTNGTSKI KMKWGMEMRR CLLQSLQQIE SMIEAESSVK EKDMTKEFFE NKSETWPIGE SPKGVEDGSI GKVCRTL LA KSVFNSLYAS PQLEGFSAES RKLLLVVQAL RDNLEPGTFD LEGLYEAIEE CLINDPWVLL NASWFNSFLT HALR UniProtKB: Polymerase acidic protein |
-Macromolecule #3: RNA-directed RNA polymerase catalytic subunit
| Macromolecule | Name: RNA-directed RNA polymerase catalytic subunit / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism: Influenza A virus (A/nt/60/1968(H3N2)) |
| Molecular weight | Theoretical: 86.524086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDVNPTLLFL KVPAQNAIST TFPYTGDPPY SHGTGTGYTM DTVNRTHQYS EKGKWTTNTE TGAPQLNPID GPLPEDNEPS GYAQTDCVL EAMAFLEESH PGIFENSCLE TMEVVQQTRV DRLTQGRQTY DWTLNRNQPA ATALANTIEV FRSNGLTANE S GRLIDFLK ...String: MDVNPTLLFL KVPAQNAIST TFPYTGDPPY SHGTGTGYTM DTVNRTHQYS EKGKWTTNTE TGAPQLNPID GPLPEDNEPS GYAQTDCVL EAMAFLEESH PGIFENSCLE TMEVVQQTRV DRLTQGRQTY DWTLNRNQPA ATALANTIEV FRSNGLTANE S GRLIDFLK DVMESMDKEE MEITTHFQRK RRVRDNMTKK MVTQRTIGKK KQRVNKRSYL IRALTLNTMT KDAERGKLKR RA IATPGMQ IRGFVYFVET LARSICEKLE QSGLPVGGNE KKAKLANVVR KMMTNSQDTE LSFTITGDNT KWNENQNPRM FLA MITYIT KNQPEWFRNV LSIAPIMFSN KMARLGKGYM FESKSMKLRT QIPAEMLASI DLKYFNESTR KKIEKIRPLL IDGT ASLSP GMMMGMFNML STVLGVSILN LGQKRYTKTT YWWDGLQSSD DFALIVNAPN HEGIQAGVDR FYRTCKLVGI NMSKK KSYI NRTGTFEFTS FFYRYGFVAN FSMELPSFGV SGINESADMS IGVTVIKNNM INNDLGPATA QMALQLFIKD YRYTYR CHR GDTQIQTRRS FELKKLWEQT RSKAGLLVSD GGPNLYNIRN LHIPEVCLKW ELMDEDYQGR LCNPLNPFVS HKEIESV NN AVVMPAHGPA KSMEYDAVAT THSWIPKRNR SILNTSQRGI LEDEQMYQKC CNLFEKFFPS SSYRRPVGIS SMVEAMVS R ARIDARIDFE SGRIKKEEFA EIMKICSTIE ELRRQK UniProtKB: RNA-directed RNA polymerase catalytic subunit |
-Macromolecule #4: Polymerase basic protein 2
| Macromolecule | Name: Polymerase basic protein 2 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Influenza A virus (A/nt/60/1968(H3N2)) |
| Molecular weight | Theoretical: 86.163391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MERIKELRNL MSQSRTREIL TKTTVDHMAI IKKYTSGRQE KNPSLRMKWM MAMKYPITAD KRITEMVPER NEQGQTLWSK MSDAGSDRV MVSPLAVTWW NRNGPMTSTV HYPKVYKTYF EKVERLKHGT FGPVHFRNQV KIRRRVDINP GHADLSAKEA Q DVIMEVVF ...String: MERIKELRNL MSQSRTREIL TKTTVDHMAI IKKYTSGRQE KNPSLRMKWM MAMKYPITAD KRITEMVPER NEQGQTLWSK MSDAGSDRV MVSPLAVTWW NRNGPMTSTV HYPKVYKTYF EKVERLKHGT FGPVHFRNQV KIRRRVDINP GHADLSAKEA Q DVIMEVVF PNEVGARILT SESQLTITKE KKEELQDCKI SPLMVAYMLE RELVRKTRFL PVAGGTSSVY IEVLHLTQGT CW EQMYTPG GEVRNDDVDQ SLIIAARNIV RRAAVSADPL ASLLEMCHST QIGGTRMVDI LRQNPTEEQA VDICKAAMGL RIS SSFSFG GFTFKRTSGS SIKREEELLT GNLQTLKIRV HDGYEEFTMV GKRATAILRK ATRRLVQLIV SGRDEQSVAE AIIV AMVFS QEDCMIKAVR GDLNFVNRAN QRLNPMHQLL RHFQKDAKVL FQNWGIEHID NVMGMIGVLP DMTPSTEMSM RGIRV SKMG VDEYSSTERV VVSIDRFLRV RDQRGNVLLS PEEVSETQGT EKLTITYSSS MMWEINGPES VLVNTYQWII RNWETV KIQ WSQNPTMLYN KMEFEPFQSL VPKAIRGQYS GFVRTLFQQM RDVLGTFDTT QIIKLLPFAA APPKQSRMQF SSLTVNV RG SGMRILVRGN SPAFNYNKTT KRLTILGKDA GTLIEDPDEG TSGVESAVLR GFLILGKEDR RYGPALSINE LSNLAKGE K ANVLIGQGDV VLVMKRKRDS SILTDSQTAT KRIRMAIN UniProtKB: Polymerase basic protein 2 |
-Macromolecule #5: RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*G)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*G)-3') / type: rna / ID: 5 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/nt/60/1968(H3N2)) Influenza A virus (A/nt/60/1968(H3N2)) |
| Molecular weight | Theoretical: 4.901097 KDa |
| Sequence | String: (P)AGCAAAAGC AGGCC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 / Component:
Details: Sample was purified in 20 mM HEPES, pH 7.5, 150 mM NaCl with Tween 20 added to a final concentration 0f 0.05% prior to plunging grids. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 3.5 sec before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2456 / Average exposure time: 6.0 sec. / Average electron dose: 1.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.7000000000000001 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Details | Rigid body fit only | ||||||||||||||
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 95 | ||||||||||||||
| Output model |  PDB-6qxe: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)