+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4452 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AL amyloid fibril from a lambda 1 light chain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | amyloid fibril / beta sheet / antibody / heart / PROTEIN FIBRIL | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
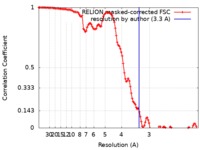
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Radamaker L / Schmidt M / Faendrich M / Fritz G | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis. Authors: Lynn Radamaker / Yin-Hsi Lin / Karthikeyan Annamalai / Stefanie Huhn / Ute Hegenbart / Stefan O Schönland / Günter Fritz / Matthias Schmidt / Marcus Fändrich /  Abstract: Amyloid fibrils derived from antibody light chains are key pathogenic agents in systemic AL amyloidosis. They can be deposited in multiple organs but cardiac amyloid is the major risk factor of ...Amyloid fibrils derived from antibody light chains are key pathogenic agents in systemic AL amyloidosis. They can be deposited in multiple organs but cardiac amyloid is the major risk factor of mortality. Here we report the structure of a λ1 AL amyloid fibril from an explanted human heart at a resolution of 3.3 Å which we determined using cryo-electron microscopy. The fibril core consists of a 91-residue segment presenting an all-beta fold with ten mutagenic changes compared to the germ line. The conformation differs substantially from natively folded light chains: a rotational switch around the intramolecular disulphide bond being the crucial structural rearrangement underlying fibril formation. Our structure provides insight into the mechanism of protein misfolding and the role of patient-specific mutations in pathogenicity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4452.map.gz emd_4452.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4452-v30.xml emd-4452-v30.xml emd-4452.xml emd-4452.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4452_fsc.xml emd_4452_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4452.png emd_4452.png | 119.5 KB | ||
| Filedesc metadata |  emd-4452.cif.gz emd-4452.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4452 http://ftp.pdbj.org/pub/emdb/structures/EMD-4452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4452 | HTTPS FTP |
-Related structure data
| Related structure data |  6ic3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10245 (Title: AL amyloid fibril from a lambda 1 light chain / Data size: 44.7 EMPIAR-10245 (Title: AL amyloid fibril from a lambda 1 light chain / Data size: 44.7 Data #1: Aligned, motion-corrected micrographs of AL fibrils extracted from human heart tissue [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4452.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4452.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.041 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Amyloid fibril of an antibody lambda 1 light chain
| Entire | Name: Amyloid fibril of an antibody lambda 1 light chain |
|---|---|
| Components |
|
-Supramolecule #1: Amyloid fibril of an antibody lambda 1 light chain
| Supramolecule | Name: Amyloid fibril of an antibody lambda 1 light chain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Extracted fibrils from the heart of a patient suffering from systemic AL amyloidosis |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: heart / Tissue: heart muscle Homo sapiens (human) / Organ: heart / Tissue: heart muscle |
| Molecular weight | Theoretical: 2.5 kDa/nm |
-Macromolecule #1: lambda 1 light chain fragment, residues 3-118
| Macromolecule | Name: lambda 1 light chain fragment, residues 3-118 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Heart / Tissue: heart muscle Homo sapiens (human) / Organ: Heart / Tissue: heart muscle |
| Molecular weight | Theoretical: 12.177463 KDa |
| Sequence | String: VLTQPPSASG TPGQRVTISC SGRSSNIGRN LVKWYQQFPG TAPKLLIYSN DQRPSGVPDR FSGSKSGTSA SLAVSGLQSE DEADYYCAA WDATLNAWVF GGGTKLTVLS QPKAAPS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Formula: H2O / Component - Name: distilled water |
|---|---|
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 294 K / Instrument: GATAN CRYOPLUNGE 3 Details: blotted from the backside for 4 s before plunging. |
| Details | Sample in pure water, pH not determined |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-30 / Number grids imaged: 1 / Number real images: 847 / Average exposure time: 6.0 sec. / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Refinement strategy included global minimization and local grid search and ADP were refined. Secondary structure restraints and NCS were applied during refinement. |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 111.4 Target criteria: REAL-SPACE (WEIGHTED MAP SUM AT ATOM CENTERS) |
| Output model |  PDB-6ic3: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)