+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | In-situ complex I with Q10 (State-beta) | |||||||||
 マップデータ マップデータ | In-situ complex I with Q10 intermediate-I | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | in-situ cryo-EM structure / mammalian / mitochondria / respiratory supercomplex / proton pumping / membrane protein / electron transport | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報: / RHOG GTPase cycle / Complex I biogenesis / Respiratory electron transport / Mitochondrial protein degradation / Neutrophil degranulation / mesenchymal stem cell proliferation / reproductive system development / mesenchymal stem cell differentiation / circulatory system development ...: / RHOG GTPase cycle / Complex I biogenesis / Respiratory electron transport / Mitochondrial protein degradation / Neutrophil degranulation / mesenchymal stem cell proliferation / reproductive system development / mesenchymal stem cell differentiation / circulatory system development / cardiac muscle tissue development / oxidoreductase activity, acting on NAD(P)H / stem cell division / ubiquinone binding / electron transport coupled proton transport / NADH:ubiquinone reductase (H+-translocating) / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / response to cAMP / NADH dehydrogenase (ubiquinone) activity / quinone binding / ATP synthesis coupled electron transport / muscle contraction / reactive oxygen species metabolic process / aerobic respiration / regulation of mitochondrial membrane potential / respiratory electron transport chain / DNA damage response, signal transduction by p53 class mediator / kidney development / fatty acid metabolic process / electron transport chain / brain development / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / multicellular organism growth / NAD binding / fatty acid biosynthetic process / cellular senescence / FMN binding / nervous system development / 4 iron, 4 sulfur cluster binding / gene expression / mitochondrial inner membrane / nuclear body / mitochondrial matrix / mitochondrion / nucleoplasm / metal ion binding 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.4 Å | |||||||||
 データ登録者 データ登録者 | Zheng W / Zhu J / Zhang K | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2024 ジャーナル: Nature / 年: 2024タイトル: High-resolution in situ structures of mammalian respiratory supercomplexes. 著者: Wan Zheng / Pengxin Chai / Jiapeng Zhu / Kai Zhang /   要旨: Mitochondria play a pivotal part in ATP energy production through oxidative phosphorylation, which occurs within the inner membrane through a series of respiratory complexes. Despite extensive in ...Mitochondria play a pivotal part in ATP energy production through oxidative phosphorylation, which occurs within the inner membrane through a series of respiratory complexes. Despite extensive in vitro structural studies, determining the atomic details of their molecular mechanisms in physiological states remains a major challenge, primarily because of loss of the native environment during purification. Here we directly image porcine mitochondria using an in situ cryo-electron microscopy approach. This enables us to determine the structures of various high-order assemblies of respiratory supercomplexes in their native states. We identify four main supercomplex organizations: IIIIIV, IIIIIV, IIIIIV and IIIIIV, which potentially expand into higher-order arrays on the inner membranes. These diverse supercomplexes are largely formed by 'protein-lipids-protein' interactions, which in turn have a substantial impact on the local geometry of the surrounding membranes. Our in situ structures also capture numerous reactive intermediates within these respiratory supercomplexes, shedding light on the dynamic processes of the ubiquinone/ubiquinol exchange mechanism in complex I and the Q-cycle in complex III. Structural comparison of supercomplexes from mitochondria treated under different conditions indicates a possible correlation between conformational states of complexes I and III, probably in response to environmental changes. By preserving the native membrane environment, our approach enables structural studies of mitochondrial respiratory supercomplexes in reaction at high resolution across multiple scales, from atomic-level details to the broader subcellular context. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_42167.map.gz emd_42167.map.gz | 62.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-42167-v30.xml emd-42167-v30.xml emd-42167.xml emd-42167.xml | 65.1 KB 65.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_42167.png emd_42167.png | 74.3 KB | ||
| Filedesc metadata |  emd-42167.cif.gz emd-42167.cif.gz | 14.2 KB | ||
| その他 |  emd_42167_half_map_1.map.gz emd_42167_half_map_1.map.gz emd_42167_half_map_2.map.gz emd_42167_half_map_2.map.gz | 116 MB 116 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42167 http://ftp.pdbj.org/pub/emdb/structures/EMD-42167 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42167 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42167 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_42167_validation.pdf.gz emd_42167_validation.pdf.gz | 1.2 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_42167_full_validation.pdf.gz emd_42167_full_validation.pdf.gz | 1.2 MB | 表示 | |
| XML形式データ |  emd_42167_validation.xml.gz emd_42167_validation.xml.gz | 13.6 KB | 表示 | |
| CIF形式データ |  emd_42167_validation.cif.gz emd_42167_validation.cif.gz | 16.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42167 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42167 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42167 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42167 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8ueqMC  8ud1C  8ueoC  8uepC  8uerC  8uesC  8uetC  8ueuC  8uevC  8uewC  8uexC  8ueyC  8uezC  8ugdC  8ugeC  8ugfC  8uggC  8ughC  8ugiC  8ugjC  8ugkC  8uglC  8ugnC  8ugpC  8ugrC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_42167.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_42167.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | In-situ complex I with Q10 intermediate-I | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||
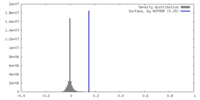
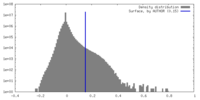
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: In-situ complex I with Q10 intermediate-I
| ファイル | emd_42167_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | In-situ complex I with Q10 intermediate-I | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
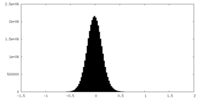
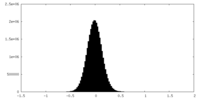
| 密度ヒストグラム |
-ハーフマップ: In-situ complex I with Q10 intermediate-I
| ファイル | emd_42167_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | In-situ complex I with Q10 intermediate-I | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
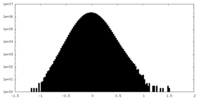
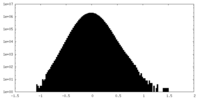
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : In-situ cryo-EM structure of respiratory supercomplex by directly...
+超分子 #1: In-situ cryo-EM structure of respiratory supercomplex by directly...
+分子 #1: NADH-ubiquinone oxidoreductase chain 3
+分子 #2: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial
+分子 #3: NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial
+分子 #4: NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial
+分子 #5: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial
+分子 #6: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
+分子 #7: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial
+分子 #8: NADH-ubiquinone oxidoreductase chain 1
+分子 #9: NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial
+分子 #10: NADH-ubiquinone oxidoreductase chain 6
+分子 #11: NADH-ubiquinone oxidoreductase chain 4L
+分子 #12: NADH-ubiquinone oxidoreductase chain 5
+分子 #13: NADH-ubiquinone oxidoreductase chain 4
+分子 #14: NADH-ubiquinone oxidoreductase chain 2
+分子 #15: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mi...
+分子 #16: NADH:ubiquinone oxidoreductase subunit A9
+分子 #17: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial
+分子 #18: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial
+分子 #19: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2
+分子 #20: NADH:ubiquinone oxidoreductase subunit AB1
+分子 #21: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 isof...
+分子 #22: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6
+分子 #23: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8
+分子 #24: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11
+分子 #25: NADH:ubiquinone oxidoreductase subunit A13
+分子 #26: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1
+分子 #27: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3
+分子 #28: NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial
+分子 #29: NADH dehydrogenase [ubiquinone] 1 subunit C2
+分子 #30: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5
+分子 #31: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 [Sus ...
+分子 #32: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mit...
+分子 #33: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mito...
+分子 #34: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6
+分子 #35: NADH:ubiquinone oxidoreductase subunit B2
+分子 #36: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3
+分子 #37: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mito...
+分子 #38: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4
+分子 #39: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9
+分子 #40: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7
+分子 #41: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10
+分子 #42: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+分子 #43: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7
+分子 #44: NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial
+分子 #45: IRON/SULFUR CLUSTER
+分子 #46: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+分子 #47: UBIQUINONE-10
+分子 #48: FE2/S2 (INORGANIC) CLUSTER
+分子 #49: FLAVIN MONONUCLEOTIDE
+分子 #50: POTASSIUM ION
+分子 #51: MYRISTIC ACID
+分子 #52: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+分子 #53: CARDIOLIPIN
+分子 #54: GUANOSINE-5'-TRIPHOSPHATE
+分子 #55: MAGNESIUM ION
+分子 #56: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+分子 #57: ZINC ION
+分子 #58: ~{S}-[2-[3-[[(2~{R})-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butan...
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 3.0 µm / 最小 デフォーカス(公称値): 1.3 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: NONE |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 3.4 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 29000 |
| 初期 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
| 最終 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
 ムービー
ムービー コントローラー
コントローラー









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)