[English] 日本語
 Yorodumi
Yorodumi- EMDB-40916: Cryo-EM structure of cinacalcet-bound human calcium-sensing recep... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Family C GPCR / Calcium-sensing Receptor (CaSR) / Heterotrimeric G protein / Cryo-EM / Lipid Nanodiscs / Positive Allosteric Modulator / Membrane Protein / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of presynaptic membrane potential / bile acid secretion / chemosensory behavior / cellular response to peptide / response to fibroblast growth factor / cellular response to vitamin D / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / negative regulation of adenylate cyclase activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / calcium ion import ...regulation of presynaptic membrane potential / bile acid secretion / chemosensory behavior / cellular response to peptide / response to fibroblast growth factor / cellular response to vitamin D / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / negative regulation of adenylate cyclase activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / calcium ion import / GTP metabolic process / positive regulation of positive chemotaxis / fat pad development / cellular response to hepatocyte growth factor stimulus / branching morphogenesis of an epithelial tube / amino acid binding / positive regulation of calcium ion import / positive regulation of macroautophagy / regulation of calcium ion transport / cellular response to low-density lipoprotein particle stimulus / anatomical structure morphogenesis / detection of calcium ion / Adenylate cyclase inhibitory pathway / JNK cascade / positive regulation of vasoconstriction / axon terminus / ossification / chloride transmembrane transport / response to ischemia / G protein-coupled receptor binding / cellular response to glucose stimulus / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor activity / positive regulation of insulin secretion / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / centriolar satellite / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / intracellular calcium ion homeostasis / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / GDP binding / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / vasodilation / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / integrin binding / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / presynaptic membrane / GTPase binding / retina development in camera-type eye / Ca2+ pathway / midbody / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / fibroblast proliferation / G alpha (i) signalling events / basolateral plasma membrane / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / cellular response to hypoxia / G alpha (q) signalling events / transmembrane transporter binding / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / positive regulation of ERK1 and ERK2 cascade / ciliary basal body / G protein-coupled receptor signaling pathway / apical plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | He F / Wu C / Gao Y / Skiniotis G | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Allosteric modulation and G-protein selectivity of the Ca-sensing receptor. Authors: Feng He / Cheng-Guo Wu / Yang Gao / Sabrina N Rahman / Magda Zaoralová / Makaía M Papasergi-Scott / Ting-Jia Gu / Michael J Robertson / Alpay B Seven / Lingjun Li / Jesper M Mathiesen / Georgios Skiniotis /    Abstract: The calcium-sensing receptor (CaSR) is a family C G-protein-coupled receptor (GPCR) that has a central role in regulating systemic calcium homeostasis. Here we use cryo-electron microscopy and ...The calcium-sensing receptor (CaSR) is a family C G-protein-coupled receptor (GPCR) that has a central role in regulating systemic calcium homeostasis. Here we use cryo-electron microscopy and functional assays to investigate the activation of human CaSR embedded in lipid nanodiscs and its coupling to functional G versus G proteins in the presence and absence of the calcimimetic drug cinacalcet. High-resolution structures show that both G and G drive additional conformational changes in the activated CaSR dimer to stabilize a more extensive asymmetric interface of the seven-transmembrane domain (7TM) that involves key protein-lipid interactions. Selective G and G coupling by the receptor is achieved through substantial rearrangements of intracellular loop 2 and the C terminus, which contribute differentially towards the binding of the two G-protein subtypes, resulting in distinct CaSR-G-protein interfaces. The structures also reveal that natural polyamines target multiple sites on CaSR to enhance receptor activation by zipping negatively charged regions between two protomers. Furthermore, we find that the amino acid L-tryptophan, a well-known ligand of CaSR extracellular domains, occupies the 7TM bundle of the G-protein-coupled protomer at the same location as cinacalcet and other allosteric modulators. Together, these results provide a framework for G-protein activation and selectivity by CaSR, as well as its allosteric modulation by endogenous and exogenous ligands. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40916.map.gz emd_40916.map.gz | 391.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40916-v30.xml emd-40916-v30.xml emd-40916.xml emd-40916.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40916.png emd_40916.png | 100.3 KB | ||
| Filedesc metadata |  emd-40916.cif.gz emd-40916.cif.gz | 7.6 KB | ||
| Others |  emd_40916_additional_1.map.gz emd_40916_additional_1.map.gz emd_40916_additional_2.map.gz emd_40916_additional_2.map.gz emd_40916_additional_3.map.gz emd_40916_additional_3.map.gz | 209.6 MB 205.9 MB 206 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40916 http://ftp.pdbj.org/pub/emdb/structures/EMD-40916 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40916 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40916 | HTTPS FTP |
-Validation report
| Summary document |  emd_40916_validation.pdf.gz emd_40916_validation.pdf.gz | 517.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40916_full_validation.pdf.gz emd_40916_full_validation.pdf.gz | 516.7 KB | Display | |
| Data in XML |  emd_40916_validation.xml.gz emd_40916_validation.xml.gz | 7.7 KB | Display | |
| Data in CIF |  emd_40916_validation.cif.gz emd_40916_validation.cif.gz | 8.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40916 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40916 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40916 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40916 | HTTPS FTP |
-Related structure data
| Related structure data |  8szhMC  8szfC  8szgC  8sziC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40916.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40916.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8677 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local unsharpened refinement map of CaSR VFT and CRD domains
| File | emd_40916_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local unsharpened refinement map of CaSR VFT and CRD domains | ||||||||||||
| Projections & Slices |
| ||||||||||||
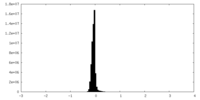
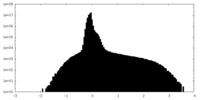
| Density Histograms |
-Additional map: Local unsharpened refinement map of CaSR CRD and 7TM domains
| File | emd_40916_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local unsharpened refinement map of CaSR CRD and 7TM domains | ||||||||||||
| Projections & Slices |
| ||||||||||||
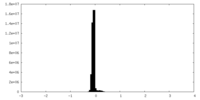
| Density Histograms |
-Additional map: Local unsharpened refinement map of Gi3
| File | emd_40916_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local unsharpened refinement map of Gi3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
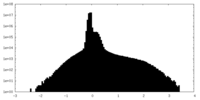
| Density Histograms |
- Sample components
Sample components
+Entire : Cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex i...
+Supramolecule #1: Cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex i...
+Macromolecule #1: Extracellular calcium-sensing receptor
+Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-3
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #6: N-[(1R)-1-(naphthalen-1-yl)ethyl]-3-[3-(trifluoromethyl)phenyl]pr...
+Macromolecule #7: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #9: CALCIUM ION
+Macromolecule #10: TRYPTOPHAN
+Macromolecule #11: PHOSPHATE ION
+Macromolecule #12: SPERMINE
+Macromolecule #13: CHOLESTEROL
+Macromolecule #14: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 262847 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)