[English] 日本語
 Yorodumi
Yorodumi- EMDB-39230: Cryo EM structure of human phosphate channel XPR1 at intermediate... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM structure of human phosphate channel XPR1 at intermediate state | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | phosphate channel / membrane protein / phosphate homeostasis / TRANSPORT PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationphosphate transmembrane transporter activity / phosphate ion transport / intracellular phosphate ion homeostasis / cellular response to phosphate starvation / phosphate ion transmembrane transport / inositol hexakisphosphate binding / efflux transmembrane transporter activity / response to virus / virus receptor activity / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
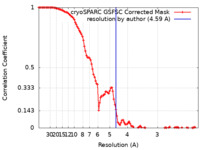
| Method | single particle reconstruction / cryo EM / Resolution: 4.59 Å | |||||||||||||||
 Authors Authors | Lu Y / Yue C / Zhang L / Yao D / Yu Y / Cao Y | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1. Authors: Yi Lu / Chen-Xi Yue / Li Zhang / Deqiang Yao / Ying Xia / Qing Zhang / Xinchen Zhang / Shaobai Li / Yafeng Shen / Mi Cao / Chang-Run Guo / An Qin / Jie Zhao / Lu Zhou / Ye Yu / Yu Cao /   Abstract: Precise regulation of intracellular phosphate (Pi) is critical for cellular function, with xenotropic and polytropic retrovirus receptor 1 (XPR1) serving as the sole Pi exporter in humans. The ...Precise regulation of intracellular phosphate (Pi) is critical for cellular function, with xenotropic and polytropic retrovirus receptor 1 (XPR1) serving as the sole Pi exporter in humans. The mechanism of Pi efflux, activated by inositol pyrophosphates (PP-IPs), has remained unclear. This study presents cryo-electron microscopy structures of XPR1 in multiple conformations, revealing a transmembrane pathway for Pi export and a dual-binding activation pattern for PP-IPs. A canonical binding site is located at the dimeric interface of Syg1/Pho81/XPR1 (SPX) domains, and a second site, biased toward PP-IPs, is found between the transmembrane and SPX domains. By integrating structural studies with electrophysiological analyses, we characterized XPR1 as an inositol phosphates (IPs)/PP-IPs-activated phosphate channel. The interplay among its transmembrane domains, SPX domains, and IPs/PP-IPs orchestrates the conformational transition between its closed and open states. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39230.map.gz emd_39230.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39230-v30.xml emd-39230-v30.xml emd-39230.xml emd-39230.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39230_fsc.xml emd_39230_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_39230.png emd_39230.png | 82.7 KB | ||
| Filedesc metadata |  emd-39230.cif.gz emd-39230.cif.gz | 5.7 KB | ||
| Others |  emd_39230_half_map_1.map.gz emd_39230_half_map_1.map.gz emd_39230_half_map_2.map.gz emd_39230_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39230 http://ftp.pdbj.org/pub/emdb/structures/EMD-39230 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39230 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39230 | HTTPS FTP |
-Validation report
| Summary document |  emd_39230_validation.pdf.gz emd_39230_validation.pdf.gz | 820.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39230_full_validation.pdf.gz emd_39230_full_validation.pdf.gz | 820 KB | Display | |
| Data in XML |  emd_39230_validation.xml.gz emd_39230_validation.xml.gz | 19 KB | Display | |
| Data in CIF |  emd_39230_validation.cif.gz emd_39230_validation.cif.gz | 24.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39230 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39230 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39230 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39230 | HTTPS FTP |
-Related structure data
| Related structure data |  8yfuMC  8yetC  8yexC  8yf4C  8yfdC  8yfwC  8yfxC  9iwsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39230.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39230.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_39230_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_39230_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo EM structure of human phosphate channel XPR1 at intermediate...
| Entire | Name: Cryo EM structure of human phosphate channel XPR1 at intermediate state |
|---|---|
| Components |
|
-Supramolecule #1: Cryo EM structure of human phosphate channel XPR1 at intermediate...
| Supramolecule | Name: Cryo EM structure of human phosphate channel XPR1 at intermediate state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 53 member 1
| Macromolecule | Name: Solute carrier family 53 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.388328 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QYEAFKDMLY SAQDQAPSVE VTDEDTVKRY FAKFEEKFFQ TCEKELAKIN TFYSEKLAEA QRRFATLQNE LQSSLDAQKE STGVTTLRQ RRKPVFHLSH EERVQHRNIK DLKLAFSEFY LSLILLQNYQ NLNFTGFRKI LKKHDKILET SRGADWRVAH V EVAPFYTC ...String: QYEAFKDMLY SAQDQAPSVE VTDEDTVKRY FAKFEEKFFQ TCEKELAKIN TFYSEKLAEA QRRFATLQNE LQSSLDAQKE STGVTTLRQ RRKPVFHLSH EERVQHRNIK DLKLAFSEFY LSLILLQNYQ NLNFTGFRKI LKKHDKILET SRGADWRVAH V EVAPFYTC KKINQLISET EAVVTNELED GDRQKAMKRL RVPPLGAAQP APAWTTFRVG LFCGIFIVLN ITLVLAAVFK LE TDRSIWP LIRIYRGGFL LIEFLFLLGI NTYGWRQAGV NHVLIFELNP RSNLSHQHLF EIAGFLGILW CLSLLACFFA PIS VIPTYV YPLALYGFMV FFLINPTKTF YYKSRFWLLK LLFRVFTAPF HKVGFADFWL ADQLNSLSVI LMDLEYMICF YSLE LKWDE SKGLLPNNSE ESGICHKYTY GVRAIVQCIP AWLRFIQCLR RYRDTKRAFP HLVNAGKYST TFFMVTFAAL YSTHK ERGH SDTMVFFYLW IVFYIISSCY TLIWDLKMDW GLFDKNAGEN TFLREEIVYP QKAYYYCAII EDVILRFAWT IQISIT STT LLPHSGDIIA TVFAPLEVFR RFVWNFFRLE NEHLNNC UniProtKB: Solute carrier family 53 member 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)