Yorodumi
Yorodumi+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3847 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the 90S pre-ribosome from Chaetomium thermophilum | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Ribosome biogenesis / RIBOSOME | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationbox H/ACA snoRNP complex / snRNA pseudouridine synthesis / tRNA cytidine N4-acetyltransferase activity / rRNA acetylation involved in maturation of SSU-rRNA / 18S rRNA cytidine N-acetyltransferase activity / tRNA acetylation / Noc4p-Nop14p complex / t-UTP complex / Pwp2p-containing subcomplex of 90S preribosome / Mpp10 complex ...box H/ACA snoRNP complex / snRNA pseudouridine synthesis / tRNA cytidine N4-acetyltransferase activity / rRNA acetylation involved in maturation of SSU-rRNA / 18S rRNA cytidine N-acetyltransferase activity / tRNA acetylation / Noc4p-Nop14p complex / t-UTP complex / Pwp2p-containing subcomplex of 90S preribosome / Mpp10 complex / rRNA (pseudouridine) methyltransferase activity / snoRNA guided rRNA 2'-O-methylation / histone H2AQ104 methyltransferase activity / box C/D sno(s)RNA 3'-end processing / rRNA methyltransferase activity / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D methylation guide snoRNP complex / positive regulation of rRNA processing / rRNA primary transcript binding / sno(s)RNA-containing ribonucleoprotein complex / rRNA base methylation / U3 snoRNA binding / snoRNA binding / positive regulation of transcription by RNA polymerase I / RNA nuclease activity / 90S preribosome / RNA processing / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / RNA endonuclease activity / RNA splicing / methyltransferase activity / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / spliceosomal complex / maturation of SSU-rRNA / small-subunit processome / mRNA splicing, via spliceosome / mRNA processing / rRNA processing / ribosome biogenesis / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / methylation / cytoplasmic translation / tRNA binding / rRNA binding / protein ubiquitination / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / GTPase activity / GTP binding / nucleolus / RNA binding / nucleoplasm / ATP binding / nucleus / cytoplasm Similarity search - Function WD repeat-containing protein 75 first beta-propeller / : / Ribosomal RNA assembly KRR1 / : / : / : / KRR1 small subunit processome component, second KH domain / Possible tRNA binding domain / RNA cytidine acetyltransferase NAT10 / Possible tRNA binding domain ...WD repeat-containing protein 75 first beta-propeller / : / Ribosomal RNA assembly KRR1 / : / : / : / KRR1 small subunit processome component, second KH domain / Possible tRNA binding domain / RNA cytidine acetyltransferase NAT10 / Possible tRNA binding domain / Helicase domain / tRNA(Met) cytidine acetyltransferase TmcA, N-terminal / TmcA/NAT10/Kre33 / RNA cytidine acetyltransferase NAT10/TcmA, helicase domain / tRNA(Met) cytidine acetyltransferase TmcA, N-terminal / GNAT acetyltransferase 2 / Sof1-like protein / : / Sof1-like domain / BING4, C-terminal domain / WD repeat-containing protein WDR46/Utp7 / BING4CT (NUC141) domain / BING4CT (NUC141) domain / U3 small nucleolar RNA-associated protein 6 / : / U3 small nucleolar RNA-associated protein 6 / U3 small nucleolar RNA-associated protein 4 / Small-subunit processome, Utp11 / Nucleolar protein 14 / Fcf2 pre-rRNA processing, C-terminal / Fcf2/DNTTIP2 / Utp11 protein / Nop14-like family / Fcf2 pre-rRNA processing / : / U3 small nucleolar RNA-associated protein 15, C-terminal / UTP15 C terminal / rRNA-processing protein Fcf1, PIN domain / WDR3 second beta-propeller domain / WDR3 first beta-propeller domain / Small-subunit processome, Utp14 / U3 small nucleolar RNA-associated protein 18 / Utp14 protein / U3 small nucleolar RNA-associated protein 13, C-terminal / Periodic tryptophan protein 2 / Utp13 specific WD40 associated domain / BP28, C-terminal domain / Nucleolar protein 58/56, N-terminal / U3 small nucleolar RNA-associated protein 10, N-terminal / U3 small nucleolar RNA-associated protein 10 / : / BP28CT (NUC211) domain / NOP5NT (NUC127) domain / U3 small nucleolar RNA-associated protein 10 / HEATR1-like, HEAT repeats / BP28CT (NUC211) domain / rRNA-processing protein Fcf1/Utp23 / Sas10 C-terminal domain / : / Fcf1 / Sas10 C-terminal domain / Small-subunit processome, Utp12 / Dip2/Utp12 Family / Small-subunit processome, Utp21 / Utp21 specific WD40 associated putative domain / WDR36/Utp21 second beta-propeller domain / WDR36/Utp21 first beta-propeller / Ribosome biogenesis protein Bms1, N-terminal / Ribosomal RNA-processing protein Rrp9-like / Sas10/Utp3/C1D / U3 small nucleolar ribonucleoprotein complex, subunit Mpp10 / RNA 3'-terminal phosphate cyclase type 2 / Sas10/Utp3/C1D family / Mpp10 protein / RNA 3'-terminal phosphate cyclase / Ribosomal biogenesis, methyltransferase, EMG1/NEP1 / RNA 3'-terminal phosphate cyclase, insert domain / RNA 3'-terminal phosphate cyclase domain / RNA 3'-terminal phosphate cyclase, insert domain superfamily / RNA 3'-terminal phosphate cyclase domain superfamily / Nucleolar protein Nop56/Nop58 / RNA 3'-terminal phosphate cyclase / EMG1/NEP1 methyltransferase / RNA 3'-terminal phosphate cyclase (RTC), insert domain / : / Pre-mRNA-splicing factor Syf1/CNRKL1 N-terminal HAT repeat / rRNA 2'-O-methyltransferase fibrillarin-like / Fibrillarin, conserved site / Fibrillarin / Fibrillarin signature. / Fibrillarin / Krr1, KH1 domain / Krr1 KH1 domain / Quinoprotein amine dehydrogenase, beta chain-like / : / : / Eukaryotic type KH-domain (KH-domain type I) / U3 snoRNP protein/Ribosome production factor 1 / Large family of predicted nucleotide-binding domains / Ribosome biogenesis protein BMS1/TSR1, C-terminal Similarity search - Domain/homology 40S ribosomal protein s23-like protein / 40S ribosomal protein S6 / 40S ribosomal protein S8 / DDB1- and CUL4-associated factor 13 / Nucleolar protein 58 / U three protein 7 / Small-subunit processome Utp12 domain-containing protein / 40S ribosomal protein S13-like protein / RNA 3'-terminal phosphate cyclase-like protein / 40S ribosomal protein s9-like protein ...40S ribosomal protein s23-like protein / 40S ribosomal protein S6 / 40S ribosomal protein S8 / DDB1- and CUL4-associated factor 13 / Nucleolar protein 58 / U three protein 7 / Small-subunit processome Utp12 domain-containing protein / 40S ribosomal protein S13-like protein / RNA 3'-terminal phosphate cyclase-like protein / 40S ribosomal protein s9-like protein / 40S ribosomal protein S4 / 40S ribosomal protein s5-like protein / RNA cytidine acetyltransferase / Nucleolar protein 56 / U3 snoRNP protein / Pre-rRNA-processing protein PNO1 / KRR1 small subunit processome component / Bms1-type G domain-containing protein / Fcf2 pre-rRNA processing C-terminal domain-containing protein / Sas10 C-terminal domain-containing protein / Uncharacterized protein / U3 small nucleolar RNA-associated protein 10 / Small ribosomal subunit protein eS1 / Uncharacterized protein / Putative U3 snoRNP protein / 40S ribosomal protein S7 / Uncharacterized protein / Putative U3 small nucleolar ribonucleoprotein protein / 40S ribosomal protein S28-like protein / Nucleolar essential protein 1 / Uncharacterized protein / 40S ribosomal protein S16-like protein / Uncharacterized protein / Uncharacterized protein / U3 small nucleolar ribonucleoprotein protein IMP3 / Nucleolar complex protein 14 / PIN domain-containing protein / U3 small nucleolar ribonucleoprotein protein IMP4 / U3 small nucleolar RNA-associated protein 15 C-terminal domain-containing protein / 40S ribosomal protein S11-like protein / U3 small nucleolar RNA-associated protein 11 / Periodic tryptophan protein 2-like protein / 40S ribosomal protein S14-like protein / Ribosomal RNA-processing protein / U3 small nucleolar RNA-associated protein 13 C-terminal domain-containing protein / H/ACA ribonucleoprotein complex subunit 2 / rRNA 2'-O-methyltransferase fibrillarin / 40S ribosomal protein S22-like protein / Small ribosomal subunit protein eS24 Similarity search - Component | |||||||||||||||
| Biological species |  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) | |||||||||||||||
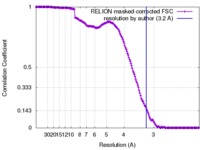
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Cheng J / Kellner N | |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2017 Journal: Nat Struct Mol Biol / Year: 2017Title: 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Authors: Jingdong Cheng / Nikola Kellner / Otto Berninghausen / Ed Hurt / Roland Beckmann /  Abstract: The 40S small ribosomal subunit is cotranscriptionally assembled in the nucleolus as part of a large chaperone complex called the 90S preribosome or small-subunit processome. Here, we present the 3.2- ...The 40S small ribosomal subunit is cotranscriptionally assembled in the nucleolus as part of a large chaperone complex called the 90S preribosome or small-subunit processome. Here, we present the 3.2-Å-resolution structure of the Chaetomium thermophilum 90S preribosome, which allowed us to build atomic structures for 34 assembly factors, including the Mpp10 complex, Bms1, Utp14 and Utp18, and the complete U3 small nucleolar ribonucleoprotein. Moreover, we visualized the U3 RNA heteroduplexes with a 5' external transcribed spacer (5' ETS) and pre-18S RNA, and their stabilization by 90S factors. Overall, the structure explains how a highly intertwined network of assembly factors and pre-rRNA guide the sequential, independent folding of the individual pre-40S domains while the RNA regions forming the 40S active sites are kept immature. Finally, by identifying the unprocessed A1 cleavage site and the nearby Utp24 endonuclease, we suggest a proofreading model for regulated 5'-ETS separation and 90S-pre-40S transition. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3847.map.gz emd_3847.map.gz | 240.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3847-v30.xml emd-3847-v30.xml emd-3847.xml emd-3847.xml | 79.4 KB 79.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3847_fsc.xml emd_3847_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3847.png emd_3847.png | 189.6 KB | ||
| Filedesc metadata |  emd-3847.cif.gz emd-3847.cif.gz | 22.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3847 http://ftp.pdbj.org/pub/emdb/structures/EMD-3847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3847 | HTTPS FTP |
-Related structure data
| Related structure data |  5oqlMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3847.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3847.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.084 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : The 90S Pre-ribosomne
| Entire | Name: The 90S Pre-ribosomne |
|---|---|
| Components |
|
+Supramolecule #1: The 90S Pre-ribosomne
| Supramolecule | Name: The 90S Pre-ribosomne / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#51 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
+Macromolecule #1: Periodic tryptophan protein 2-like protein
| Macromolecule | Name: Periodic tryptophan protein 2-like protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 100.817508 KDa |
| Sequence | String: MKTNFKFSNL LGTVYCRGNL LFSPDGTHLF SPVGNRVTVF NLVENKSYTF PFAHRKNISR IGLTPQGNLL LSIDEDGQAI LTNVPRRVV LYHFSFKSPV TALAFSPSGR HFVVGLKRKI EVWHVPSTPD TNEDGDLEFA PFVRHHTHMQ HFDDVRHLEW S SDSRFFLS ...String: MKTNFKFSNL LGTVYCRGNL LFSPDGTHLF SPVGNRVTVF NLVENKSYTF PFAHRKNISR IGLTPQGNLL LSIDEDGQAI LTNVPRRVV LYHFSFKSPV TALAFSPSGR HFVVGLKRKI EVWHVPSTPD TNEDGDLEFA PFVRHHTHMQ HFDDVRHLEW S SDSRFFLS ASKDLTARIW SLDTEEGFVP TVLSGHRQGV VGAYFSKDQE TIYTVSKDGA VFEWKYVAKP GHEDDEMVDD ED MQWRIVN KHFFMQNAAT LRCAAYHAES NLLVAGFSNG IFGLYEMPDF NLIHTLSISQ NEIDFVTINK SGEWLAFGAS KLG QLLVWE WQSESYILKQ QGHFDAMNSL VYSPDGQRIV TAADDGKIKV WDVESGFCIV TFTEHTSGVT ACEFAKKGSV LFTA SLDGS VRAWDLIRYR NFRTFTAPER LSFTCMAVDP SGEVIAAGSI DSFDIHIWSV QTGQLLDRLS GHEGPVSSLA FAPDG SVLV SGSWDRTARI WSIFSRTQTS EPLQLQSDVL DVAFRPDSKQ IAISTLDGQL TFWSVSEAQQ VSGVDGRRDV SGGRRI TDR RTAANVAGTK NFNTIRYSMD GTCLLAGGNS KYICLYSTTT MVLLKKFTVS VNLSLSGTQE FLNSKLMTEA GPVGLLD DQ GEASDLEDRI DRSLPGSKRG DPGARKKFPE VRVSGVAFSP TGNSFCAAST EGLLVYSLDN TVQFDPFDLN MEITPAST L AVLEKEKDYL KALVMAFRLN EAGLITRVYQ AIPYTDIGLV VEQFPTVYVP RLLRFVAAQT EQSPHMEFCL LWIRALIDK HGPWLAANRG KVDVELRVVA RAVAKMRDEI RRLADENVYM VDYLLNQAKS KSNNRAVVGD AEEEWGGLDD FGVKALPSSK PETTLDDIM QDDVAESEEE WIGLE UniProtKB: Periodic tryptophan protein 2-like protein |
+Macromolecule #2: Utp2
| Macromolecule | Name: Utp2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 103.960703 KDa |
| Sequence | String: MAGSQLKRLK ASLREQGLIG PQKSKKQKRQ NANDPKAQEK RLQKAEALKS IREQFNPFQF KTNARGPKFE VTTNKPTDKN GMVIKGRPE LSRARSEEKR RQTILVEMQR RNKVGGIIDR RFGEGDPNLS VEDKMIERYT QEALRAHKKK GMFDLEDDDE A RLTHMGKP ...String: MAGSQLKRLK ASLREQGLIG PQKSKKQKRQ NANDPKAQEK RLQKAEALKS IREQFNPFQF KTNARGPKFE VTTNKPTDKN GMVIKGRPE LSRARSEEKR RQTILVEMQR RNKVGGIIDR RFGEGDPNLS VEDKMIERYT QEALRAHKKK GMFDLEDDDE A RLTHMGKP LFDDDEAPKE DFSEDDLPSG DESDTTRAER RALKRQRLAE AMEGLEDEAG QPERKKTRKE IYEEIIAKSK MY KAERQAI KEENNELRME VDQELPEVQQ LLYQIKKPET EKKEAPVPII AGKEKTVVDK EYDIRLKQLA MDKRAQPAEP VKT EEEKAA EAAKKLQELE EKRLKRMRGE PVSDDEESES EEDKGKKDKK DNAVDPLNAE ESEDEFGLGK GVKYKPTATE LGLD DEDDF LIDDDLVASG SELEFDSEDF SDVESDEEEG SSESKEDEED DEFIKGILTE AEQQEAVFQQ PKQGDDENGI PYTFP CPQS HEEMLNVVKG IEVTKLPTVV QRIRALHHPK LDAKNKERLG NFSQVLIHHI AYLGDRFQPH WFPTLEQLSR HVHSLA KTF PIEVAKAYRM RIQEMEEHRP LAPTVGDLVI LTAIGTTFPT SDHFHQVCTP AMLAIARYLG QKVPAALSDF AVGIYLS IL ALQYQDFAKR YVPEMMNFLL NTLCALAPER AKSKLGNFPV HEPPAGIRIK DATNTPIRQL NCGDCLRKDE LSPAETSS L QIAILSTATA ILKSAADTWH KLPAFIESFQ PALSVAQHLL TKPNASHLPS SLTSKLNDLA SHLSRLLQLS RLSRRPLEL HHHRPLAIKT YIPKFEDDFD PDKHYDPNRE RAELAKLKAE HKRERKGALR ELRKDAQFIR REQLRIKKEK DEAYEKKFKR IIAEIQNEE GRAANEYARE KAARKRKR UniProtKB: Nucleolar complex protein 14 |
+Macromolecule #3: Utp3
| Macromolecule | Name: Utp3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 73.214633 KDa |
| Sequence | String: MAKKRKATRP AEPQGPKEID PKDARLTVNN YMDVADSEDE YWYKKDRLDI DSDDEPRSKR LKRQDKEDAF LEQSDEEIFD DELSSEEEE EDDEDERRAP TKKDGKKGAA LDDDDDAELL GLGKKKDDDE IRDEGWWGSS KKEYYNADAI ETEQDALEEE K EARRLQAK ...String: MAKKRKATRP AEPQGPKEID PKDARLTVNN YMDVADSEDE YWYKKDRLDI DSDDEPRSKR LKRQDKEDAF LEQSDEEIFD DELSSEEEE EDDEDERRAP TKKDGKKGAA LDDDDDAELL GLGKKKDDDE IRDEGWWGSS KKEYYNADAI ETEQDALEEE K EARRLQAK KLAKMQEADF AFDEAEWLAT KDEKTGGDEE VVTEVLKEVE VTSDMGPEER YNLLQARYPE FDYLVEEFRE LR PLLETLQ KEAEGKPAKS LPVVKAWVLG CYVAALASYF AILTSPTRDG DESKGIMNPS ELRDHEVMQT LMECREAWLK VKS LRPAKG AVSAHGMLSP PEEGEEAGSD VDMLDDHAVQ KPKKAKKLSK EEIKANKKKK EEEAKKAKAV EQSLAELTSL LQTT KKAAV KASVASKAMS TAATSGADSG MDDNRSDFGE EEELDARTAA EKAKRKKSLR FYTSQIVQKA NKRQGAGRDA GGDMD IPYR ERLKDRQARL NAEAERRGKR DSKFGADLGG DSSDDEDAKA RQVRDDEDAY YNEVVQAAAK KRADKQARFE ALAAAR KGD RVVEEETIGP DGKRQITWQI QKNKGLTPNR KKEQRNPRVK KRKKYEEKQK KLRSVKAVYK GGEGPGGYQG ELSGIKT NL VKSVKL UniProtKB: Sas10 C-terminal domain-containing protein |
+Macromolecule #4: Utp4
| Macromolecule | Name: Utp4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 97.696523 KDa |
| Sequence | String: MDIHRCRFVR YPASAINAVA FTHSALPVVS SSKKYLQKNI QVRLAIGRAN GDIEIWNPLN GGWYQEVIIP GGKDRSVDGL VWVTDPDEE MADGKIIHGK SRLFSIGYTT TITEWDLEKA RAKKHASGQH GEIWCFGVQP LPHKANAAAA QNRKLVAGTV D GNLVLYSI ...String: MDIHRCRFVR YPASAINAVA FTHSALPVVS SSKKYLQKNI QVRLAIGRAN GDIEIWNPLN GGWYQEVIIP GGKDRSVDGL VWVTDPDEE MADGKIIHGK SRLFSIGYTT TITEWDLEKA RAKKHASGQH GEIWCFGVQP LPHKANAAAA QNRKLVAGTV D GNLVLYSI EDGDLKFQKT LTRTPSKKTK FVSIAFQSHN IVIVGCSNST ICAYDVRTGT MLRQMTLGTD LTGGSKNIIV WA VKCLPNG DIVSGDSTGQ VCIWDGKTYT QAQRIQSHTQ DVLCLSVSAD GSKIISGGMD RRTAVYEPMA GQSGRWSKVF HRR YHQHDV KAMASFEGKG MSVVVSGGSD ASPIVLPLRA LGKEFHRTLP HLPQHPTVLS APKARYILSW WENEIRIWHL LNSA QQFLD DPQAPLNLRK NRKFLAQVLI KGASHITSAS ISEDGTLLAA STPTDVKVFH LDPAAAQRNG QLYIKKVNMT GTGLG ATRV QISPDKRWIC WAEEGSKVMI SRVHATESAD GISYTVSVPH KLHRLRRQIP KHILLGGLGS YDRNVSQIAF SADSRM LSV ADLAGYIDTW VLRGPGEGVN GTGGEDSDGE SAASSSDSSD EKSEDIAGER WARNPKAAMI PKLSAAPVVL SFSPTPR DD GDYDLLVVTT LKQLLIFNPL RGMLSEWSRR NTYPKLPEPF RDTRDQVKGI VWQGQRAWFY GVASLFMFDL SQDFSPEK D LVETNGHKQG TKRKRGAHES GAGSKIEKHS LVPQRIRAAS APDGTKWEDI EMVDADDQKS VGVSSGVDDD DDETDGSEL QRLREENREA NSSANAEKEG PSRAKWWHTY QFRPIMGIVP IEGMMEKKLG AVEGIPPLEV ALIERPLSED DLPERYFAEG EWER UniProtKB: Uncharacterized protein |
+Macromolecule #5: Utp6
| Macromolecule | Name: Utp6 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 46.924773 KDa |
| Sequence | String: MAWVADKARF YLERAAPELR EWEEKEIFTK DEIRNLVAKR SDFEHLVLAP GTKPTDFLNY VNWERSLDRL RAKRCARLNI RSVTSHASQ ARTFGIFERA VLKHPGSIEL WLAYLEFAAQ VKATKRWRRI MTRALRLHPM NASLWTLAGR RAAQNGDMQR A RAHFLRGC ...String: MAWVADKARF YLERAAPELR EWEEKEIFTK DEIRNLVAKR SDFEHLVLAP GTKPTDFLNY VNWERSLDRL RAKRCARLNI RSVTSHASQ ARTFGIFERA VLKHPGSIEL WLAYLEFAAQ VKATKRWRRI MTRALRLHPM NASLWTLAGR RAAQNGDMQR A RAHFLRGC RFCTREPTLW LEYARCEMDW LARMEAKKQG QGVRKGVNAL EAIKATEGQE EGDIIPIGED TEDDSGDEDG LI LPDPDAE GTDGTKKAAK PVFDAEQTKK LEQSPALSGA IPIAVFDVAR KQPFWGPAAA EKFFDVFAKF GHLSCHERII SHV VTTMQE LFPNHPCTWS VHIRQPLVGV DVLTPAFPKA LRESLARLKA ALQSTTDRKA LATKMVAWMD GILAIEKLDA AIRT VLEHT KRSLEESPS UniProtKB: U3 snoRNP protein |
+Macromolecule #6: Utp7
| Macromolecule | Name: Utp7 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 63.629379 KDa |
| Sequence | String: MDTSEAVDLA PAPAKQGRQA NGAHSIANAR TEFRSKAELD RIRRYKQAQK KYGRGPRVDI KSVRDKKLRR TLTNLENKYK TAALKAKEA EILLENQTGF LEPEGELERT YKVRQDEIVK EVAVEVAQKK FELKLTELGP YTCEYSRNGR DLILAGRKGH V ATMDWREG ...String: MDTSEAVDLA PAPAKQGRQA NGAHSIANAR TEFRSKAELD RIRRYKQAQK KYGRGPRVDI KSVRDKKLRR TLTNLENKYK TAALKAKEA EILLENQTGF LEPEGELERT YKVRQDEIVK EVAVEVAQKK FELKLTELGP YTCEYSRNGR DLILAGRKGH V ATMDWREG KLGCELQLGE TVRDARFLHN NQFFAVAQKK YVYIYDHNGV EIHCLRKHVE VSHMEFLPYH FLLATLSISG QL KYQDTST GQIVAEIATK HGTPVSLTQN PYNAILHIGQ QNGTVTLWSP NSTDPLVKLL AHRGPVRSLA VDREGRYMVS TGQ DNKMCI WDIRNFKEAV NSYFTRAPAT SVAISDTGLT AVGWGTHTTI WKGLFNKERP VQVKVDSPYM TWGGQGQVVE RVRW CPFED ILGIGHNEGF SSIIVPGAGE ANYDALEVNP FETKKQRQEG EVKALLNKLQ PEMIALDPNF IGNLDLRSEK QRQAE RDLD QPAQDIVEEL RKKARGRNTA LKKYLRKQRK KNIIDEKRLK AEELYRQMQE KRDGQTKEKQ AEVGPALARF VRKE UniProtKB: U three protein 7 |
+Macromolecule #7: Utp10
| Macromolecule | Name: Utp10 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 198.508438 KDa |
| Sequence | String: MASSLAQQLA QIAANSRSSF NVKALKASHS KSLIWEPRVA VSQTFAEIYS QCYEGFKELC HLDSRFVPFD ATLFSAQSQE VDRTQMTAE ENAALDKRVD SFLHLVGSRL RLMPAIKAVE WLIRRFRIHE FNTGTLLATF LPYHTIPAFV TLLSILPVQR I PIEYRFLD ...String: MASSLAQQLA QIAANSRSSF NVKALKASHS KSLIWEPRVA VSQTFAEIYS QCYEGFKELC HLDSRFVPFD ATLFSAQSQE VDRTQMTAE ENAALDKRVD SFLHLVGSRL RLMPAIKAVE WLIRRFRIHE FNTGTLLATF LPYHTIPAFV TLLSILPVQR I PIEYRFLD PYIKSLTPPP RAAIVQQATN RPDLLSAISR YTLDSCRAKQ EYPGLISFWG GIMAEAVNGM IDKMRSGRRA IQ LENDHLL LQQIGPVLSE AMVMKDVPGI QIASYMVVAI LAAKGSLNDN ILTAFMEQLV HGWTVDTLRP GLVCLTMLAQ HRS AKQLSG RVAKAVIKVP DLVSSLRDIS KEHQVDKLAN GLVLAFVDRL AKKGDIRTLP VINSLLLSEL LQEKQAKVAY KALL LAAHK IDDNVDADGN IRKQVGSALV RLSQAEGDVG DAIRTAIQEV DFNIEELELK LGAAIRPKLA IEEAPEPSDE AMTVR PVDQ RPSLDSTFER LSKLQPTATS CLAKDSESLF NDLCSVFLSA AVSESDLERF DATPVLSRPK APSNSFYLSF YLRVWC GPY PTLAKVAALE RVKTRLKEGD CVDKDFQAIF PYCIAALSDP AKKVRRAAAD LVAVLGSAYK SLEKSQQLWA AKDLYGK TG TTSPLDKDAL KALIRSVLIP CLEESVLHED HVVAALVGAL ESSKDSENKN ADKRHLSHSA RLSIFKFLCG HVVETPLL A VKLRLLRSLN QIRRISGTTR TDLLLPLLRW WAGLSANEAA ELAAQESVDV PAIDDAVVDV VVPNHAAGLE AFFQLVKEA IRPGLLQAIF ARIAKMWPSM KSDTKYSTAK TLFELTQDPK LNAEQSDVIT EAVEVLRKVD LTTDILHYFI DSLQDEVRLA TEGPANKRR RVSTTEPGRG VGTQSSPELQ AALSKTTFVL ELVQESTPAN HPELLPSLFT TLSDLHHLSA IVGSELGYLQ N LVLSSLLA MMPTYKDNKD LTIDASVGHG DVLATCIQKS TSPAVINSAL LLVASLARTA PDVVLQSVMP IFTFMGSSVL KQ ADDYSAH VVNQTIKEVI PPLIETFRKR GRNVVASAKD LLASFVTAYE HIPSHRKHNL FISLIQNLGP DDFLFAILAM FVD RYGATD NMITFMTQLI SSFTVETQLQ TLLKFLDLVG DLFKSKPTLS NVLLGGGSGL NGEQDIQKAA TKQLNLLPHL LSNR RLKRE IMQLAERDDM EAGKIRELYA ALLEGILTVA STVKTKKTLH SRCGDALSNL LNLLSIAEFI KSVEALLDRP NIGLR QKVL RALELRVDSE SSVDPKSREA LLAFLPQLTA VIRESDDMNY KHTAVICVDK ISEKYGKKDL DAVAAAATTI AGDYCL GQP SQSLRVMALL CLASLVDVLQ DGIVPVLPVA IPKALSYLEQ SVAEGGRDVE LHNAAYAFMT ALAQHIPYMI TGNYLDR LL ACSNASAAAN LDDEANANRM QCLQFLAKLV DARVLYTGLH QNWASAAKFG FSAISEYLQI LGIALDKHSK TVVVKNVS S LSSIFLSAMD LRRTVAAGDI ASQISAMELD EIETKTHEDA LKMVYKLNDA AFRPIFSKFV EWATTGLPKS DVTGRTYRL YVVFGFLDAF FGSLKSIVTG YASYIVDASV KALKAVDFAV PEERNLWKRV LCTLAKCFEH DQDGFWQAPA HFGAVAPVLV EQFLRAEGQ VTATNVNDVI QDVVPAVVEL AAAVESQEHY KEINTALLKH LRNGSPGVRL AVVKCQQAIT AKLGEDWLHL L PEMLPYIS ELQDDDDEVV ERENRRWIVG IEEKLGESLD SMLQ UniProtKB: U3 small nucleolar RNA-associated protein 10 |
+Macromolecule #8: U3 small nucleolar RNA-associated protein 11
| Macromolecule | Name: U3 small nucleolar RNA-associated protein 11 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 31.37965 KDa |
| Sequence | String: MSSLRHAIQR RAHKERAQPL ERQRLGILEK KKDYRLRARD YKKKQAVLKS LRQKAAERNE DEFYFGMMSR KGPGSALTRG KGFTGTVDG DRGNKALSVE TVRLLKTQDL GYVRTMRNIA AKELKELEER YVLAGGADQP VEEFNSDEDE EESGSKQAKP K KIVFFEGV ...String: MSSLRHAIQR RAHKERAQPL ERQRLGILEK KKDYRLRARD YKKKQAVLKS LRQKAAERNE DEFYFGMMSR KGPGSALTRG KGFTGTVDG DRGNKALSVE TVRLLKTQDL GYVRTMRNIA AKELKELEER YVLAGGADQP VEEFNSDEDE EESGSKQAKP K KIVFFEGV EERQQALEKQ KADEEMKDYD EEEDGYDFDD EEEMTEKEKE ERRKQLVLEK LARKVKAARK KLKALADAEY EL ELQQAKM AKTATSGGFT KSGRRIKVRE RKR UniProtKB: U3 small nucleolar RNA-associated protein 11 |
+Macromolecule #9: Utp12
| Macromolecule | Name: Utp12 / type: protein_or_peptide / ID: 9 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 107.573039 KDa |
| Sequence | String: MVKSYLKFEP SKSFGVVVSS NSNLVWSSRG KAGAGAGQAI VAANEEVLVW DIKKGELLSR WRDENCKFRV TAIAQSRTDP DVFAVGYED GSIRLWDSKI ATSVVSFNGH KSAITVLAFD KTGVRLASGS KDTDIIVWDL VAEVGQYKLR GHKDQVTGLY F IEPDPVVR ...String: MVKSYLKFEP SKSFGVVVSS NSNLVWSSRG KAGAGAGQAI VAANEEVLVW DIKKGELLSR WRDENCKFRV TAIAQSRTDP DVFAVGYED GSIRLWDSKI ATSVVSFNGH KSAITVLAFD KTGVRLASGS KDTDIIVWDL VAEVGQYKLR GHKDQVTGLY F IEPDPVVR TEGEDDHAVM AVDSEPSDGF LLTTGKDSLI KLWDLSSRHC IETHVAQANG ECWALGVSPD FTGCITAGND GE ITVWALD ADALASSAQK VDLSQSVNFL QNRGTLHRSS KERAAEVVFH PLRDYFAVHG VEKSVEIWRI RNEAEIKKTL ARK KKRRRE KLKEKKAEGA DADMDETDDT DIAKAEVSDV FVQHVIVRTT GKVRSVDWAL NPGVKDLQLL VGGTNNLLEL YNIV GKERL KSKGEEPDYN KALAVELPGH RTDIRSVAIS SDDKMLASAA NGSLKIWNIK TQQCIRTFDC GYALCCAFLP GDKVV IVGT KEGELQLYDV ASASLLETVN AHDGHAVWAL QVHPDGRSVV SGGADKTAKF WDFKIVQEPV LGTTRTTHRL KLVQSR ILK VSDDILSLRF SPDARLLAVA LLDSTVKVFF NDTLKLYLNL YGHKLPVLSM DISYDSKLIV TSSADKNIRI WGLDFGD CH KALFGHQDSI LQVAFIPHNS DGNGHYFFSA SKDRTIKYWD ADKFEQIQRI DGHHGEIWAL AVSHSGRFLV SASHDKSI R VWEETDEQIF LEEEREKELE ELYEQTLTTS LEQDPDEQDA NREIAAATKQ TVETLMAGER IAEALELGMT DLNTIREWE EARQINPNIA PPQRNPIFVA LGNIPAETYV LNTLQKIKPA SLHDALLVLP FSTIPSLLTF LNLFAQRELN VPLTCRILFF VLKTHHKQI VASRTMRATL EKVRANLRAA LRRQKDEMGF NIAALKVVSM QLRDKSVREY VDETWEEKEK EKGVRKRAFA S LS UniProtKB: Small-subunit processome Utp12 domain-containing protein |
+Macromolecule #10: Utp13
| Macromolecule | Name: Utp13 / type: protein_or_peptide / ID: 10 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 100.430352 KDa |
| Sequence | String: MATKQPAKTT FEVANVIQPI YTGGSVALEN GARILASTLG ENAILTELNT GKRLAEIQGD GEPISTLTIT PSGSHLIVCS RSLTMRIYS LAISPDYDSV EPTLVRTTKP HATPVVVLAV DRTSTLLATG AADGAIKIWD IIGGYVTHTV SGPSVLVSAL H FFEIAVTA ...String: MATKQPAKTT FEVANVIQPI YTGGSVALEN GARILASTLG ENAILTELNT GKRLAEIQGD GEPISTLTIT PSGSHLIVCS RSLTMRIYS LAISPDYDSV EPTLVRTTKP HATPVVVLAV DRTSTLLATG AADGAIKIWD IIGGYVTHTV SGPSVLVSAL H FFEIAVTA ESQSSNKKPK KGSRKGQNDD ADEIASRFRL AWGTQDGRIR IFDLYKRTTT PVYADPKRKK EAHESNVQCI AY SPEQHAL LSGSRDKTMT LWLWRDGIWQ GTPMLRHELL ESVGFLNEGK WMYSAGTSGV LRIWDTTTHH EITKKQDAKS EGE AILSAV SLPERSLILC AQADFTLVLY RVPSPADVVS SSEGILLEPF RRISGTHDEI LDLTYILPDQ SMMAIATNSE DIRI VSVKD AQAYSEDNAE CRSGSYFGHD VALLKGHEDI VMSLDVDWSG HWIASGSKDN TARLWRVDPA NNSYTCYAVF TGHLE SVGA VALPKVVPPA NSEAFKNPLD HPPAFLISGS QDRFVQKRDI PRQLQKGGKL TSSLRRLAHD KDINALDISP NGKLFA SAS QDKTVKIWDV EKLEVQGILR GHKRGVWTVR FAPLNTPVIQ GEQGSVSGRG VVLTGSGDKT IKLWNLSDYT CIRTFEG HS HNVLKVVWLH ISRDDSITKT KVQFASAGAD TLVKVWDANT GETECTLDNH EDRLWTLAVH SKTNILASAG SDSKVTFW R DTTAETQAAA AQAALKLVEQ EQTLENYIHA GAYRDAIVLA LQLNHPGRLL NLFTNVVTTR NPDPDSLTGL KAVDDVLAK LSDEQIFQLL LRLRDWNTNA RTAPVAQRVL WALFKSHPAN KLSSLSVKGA RGHKSLNEVL DAIKVYTERH YKRIEELVDE SYLVEYTLR EMDALTPQTE ALEAGEDAVI AEA UniProtKB: U3 small nucleolar RNA-associated protein 13 C-terminal domain-containing protein |
+Macromolecule #11: Utp14
| Macromolecule | Name: Utp14 / type: protein_or_peptide / ID: 11 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 104.741391 KDa |
| Sequence | String: MPGRQAHGRP LLPSLGSSSG GGKSKKSRSK ARRSAQSKAL DAFALAAEQV PESRSKGVRL RDLDEHPSTS ASRNKRQHED DEEDDFDGF DDDEGMGRKK RARRDSGGDE DGDDVDEDME GGADGEDDSE EWHVGVTGAD EDSEIDSDEA FGESDEERFE G FVFGGSKS ...String: MPGRQAHGRP LLPSLGSSSG GGKSKKSRSK ARRSAQSKAL DAFALAAEQV PESRSKGVRL RDLDEHPSTS ASRNKRQHED DEEDDFDGF DDDEGMGRKK RARRDSGGDE DGDDVDEDME GGADGEDDSE EWHVGVTGAD EDSEIDSDEA FGESDEERFE G FVFGGSKS HKKGTKKRRN GSEDDAIDLA TALDQYESSE EGEEGEESES EEDDEESEEE DSDEEDDDPS KLDALQSMIA GF AGEDEES GGEGEAKPSG KQKLSLKDLG LVGVKDPDMK RSLRLMKKEE KATKPGSSKK LEVPLAKRQQ DRLMREAAYK KTN ETLDRW IETVKHNRRA EHLVFPLAQN AHDRGLDASE LQPITQKTSG TELEQTILAI MEESGLGPTA KPEKKETSGE AGKQ GMSKE EQKELIRQKR RERELHSREM ARLKRIKKIK SKTWRRIHRK ELLKNEEAAY QAALEAGELD SEEEREALDR RRALE RVGA RHKESKWAKL GKKAGRAVWD EDFRAGLAEM ARRKDELRKR IEGLKNGSDD SSDDSDGDED ASGDEAAERR KLLAEL EKA AAYSDDDEPH SKLFQLKFMQ RGEELRRKEN EELIAQIRRE LDSDYDGSEP DEVEIGRRTF GMGKKDKTEQ QQAQDQL AR MQAAEARKEK AAARVAAAEE TVKQEAREKA EKDEASAPST AGAWSQPKVE AGAWSKPDDE GSSKPRKAPK GRPEEIDV T DLALAGKPAK AKANGQAKSQ VQGKAHASNG QSSSSQKAST SVAITGAEED GNDSDDSDAV HLPMAIRDRA LIERAFAGD DVAAEFEKEK AEIEADDDEK EIDNTLPGWG SWVGEGVSNR EKRRHQGRFV TKVEGIKKEK RKDYKLKDVI INEKRVKKND KYLATGLPH PFESQQQYER SLRLPVGPEW MTKETFQDAT KPRVIIKQGI IAPMSKPMY UniProtKB: Uncharacterized protein |
+Macromolecule #12: Utp15
| Macromolecule | Name: Utp15 / type: protein_or_peptide / ID: 12 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 60.524145 KDa |
| Sequence | String: MAAEVVPLPQ LKLPSGPSPI TAEQRYWRSF KKQKSHTSTA NWPISHISFP ASLGTTALVS SSLVAAAKTN DLFAVTAGPR VEIFSIRKR EPLKTIGRFD SEAHCGEIRP DGRVLVAGED TGRMQVFDVG QGTRAVILKT WHIHKQPVWV TKWSPTELTT L MSCSDDKT ...String: MAAEVVPLPQ LKLPSGPSPI TAEQRYWRSF KKQKSHTSTA NWPISHISFP ASLGTTALVS SSLVAAAKTN DLFAVTAGPR VEIFSIRKR EPLKTIGRFD SEAHCGEIRP DGRVLVAGED TGRMQVFDVG QGTRAVILKT WHIHKQPVWV TKWSPTELTT L MSCSDDKT VRLWDLPSND PTRLFTGHTD YVRCGAFMPG SANSNLLVSG SYDETVRVWD ARAPGGAVMT FKHADPIEDV LP LPSGTTL LAASGNAISV LDLVAAKPLR LITNHQKTVT SLSLASQGRR VVSGSLDGHV KVFETTSWNV VAGAKYPSPI LSL SVITAG ASHDDRHLAV GMQSGVLSIR TRLSGPAADR ERERERVEAA MAKGPEAIAK LDAAKAKRKR AAVSNKNMDL LGES ADVII PTADPGTHPR GRRPKLKPWQ KAFRQGRYAA AVDDVLNTTA PSYDPVIALT LLTALRHRSA LREALQGRDE LSVIN ILRW AGKYVADPRY RSICVDVAFH LIDLYAEHVG GSAELATQFQ QLLAKVNREV EKAELAIVTG GMVESLMMSV EAQ UniProtKB: U3 small nucleolar RNA-associated protein 15 C-terminal domain-containing protein |
+Macromolecule #13: Utp17
| Macromolecule | Name: Utp17 / type: protein_or_peptide / ID: 13 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 105.039695 KDa |
| Sequence | String: MVSTMETNGD TASLKRKREP KDDPHSLQKK HRHRSRSKPQ DAATADSVAN LNSNSLAVQP IDGNSGPLQL ATTDRLASWK VSKPMGGRM SDIDPIFSQD ERHLIITYNT SIQVYSTEDS LLVRRIALPL TRTNDLDEPS ATHIVSSALS KSNPDYLWVA C SDGRIWHI ...String: MVSTMETNGD TASLKRKREP KDDPHSLQKK HRHRSRSKPQ DAATADSVAN LNSNSLAVQP IDGNSGPLQL ATTDRLASWK VSKPMGGRM SDIDPIFSQD ERHLIITYNT SIQVYSTEDS LLVRRIALPL TRTNDLDEPS ATHIVSSALS KSNPDYLWVA C SDGRIWHI NWTSGEGVDT PSTIDTKKLL DMAVDAIEVA GKVDDVLLTL NRLTKSSAQI IAYNSKMLAT KTGKLLHTYD ES PQSLRSV AGGRAIVAAA KEALHIGILK TKKLASWEEL AYRFVSFDVP DIISTFDIRP IIRMIKKGVA ELQDIDVAVG GAR GAIYVY SNLLAHLHTE ASGPLRVGTI QPRKYHWHRR AVHSVKWSGD GNYLISGGYE TVLVLWQLDT GRVDFLPHLS AAIE NIVVS PKGSAYALHL DDNSAMVLST AEMKPSMYVS GIQSLVLGDR PSKDALVRRV WRPIDEIASP LVATISPQNP SHMFL CVGN GQQATVGGGA TSTPLVQVFD ISSFQGVAKQ AIARTNPTDV NITSEGVPII EPTATKLAFS HDGKWLASID EWQPPE RDT EAYLTGSKTQ SDACKERREI YLKFWEVGAD QSLELVTRIN DAHYTKQTES IFDLASDPTS ARFATIGNDG MVRFWSP KL RKRDGLMATR PDGQPLRSWS CSRVVPLPVH ERQDDSVEIL KGIPYSGAIT FSEDGSILFA AFGPPSGALV VAIDTQTG T VRDVVSGMFK GDIRAMKSLS SCLIMLSDDL VVYDIVSDEM LASYTLKETS EAAKKLTQLA VNHQSRSFAL AAPIPNLGQ DKLKRGTKSE LLIFNIEDEE PKLVKTFSQV IISVCAVPSS SGFVVVDSAA QVWSITEGAE QAPLLKSLAD LGVDNTSNTE MEDTPDKQL LLEQAEANDD EMHDANYDLD MEDNEDTHAV VVAPQRLAEI FNAAPAFAMP PIEDVFYQVA SLFSTKPVIN A UniProtKB: Uncharacterized protein |
+Macromolecule #14: Utp18
| Macromolecule | Name: Utp18 / type: protein_or_peptide / ID: 14 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 69.055141 KDa |
| Sequence | String: MPRSNKKKQI EDLPSDSDHF PEFPEEEAFS SLSEEEEAEE RKKPVAEKDA EEEELEKFVL GAKETFREQL FRDDFLAPAN DPTALVKAG DDEEETGHEH LDDSMLFVID TEGDSSIQVP APTKVTAEET GGDAPAWEDS DDERLTISLA GATRLRKLRI S ESEDLVSG ...String: MPRSNKKKQI EDLPSDSDHF PEFPEEEAFS SLSEEEEAEE RKKPVAEKDA EEEELEKFVL GAKETFREQL FRDDFLAPAN DPTALVKAG DDEEETGHEH LDDSMLFVID TEGDSSIQVP APTKVTAEET GGDAPAWEDS DDERLTISLA GATRLRKLRI S ESEDLVSG IEYARRLRQQ YLRLYPQPDW AKEANGSKRR RRRSLDASSD SLSGSDMEVD SDGESIDAPL PLDSFLRDAN SF KVAAEDS ARSAKRRKLR PETIDIQRTR DIPDTHKAAI SSLAFHPRYP ILLSSSTSSI MYLHKLDASA YPTPNPLLTS VHV KRTDLR RAAFVGPDGG EIIFAGRRRY FHCWNLSSGL VKKVSKIQGH QKEQRTMERF RVSPCGRYMA LVASDKKGGG MLNI INVGT MQWIAQARID GRHGVADFAW WSDGNGLTIA GRDGQVTEWS MITRRTVGIW RDEGSIGGTV MALGGRNGPA ELGGD RWVA IGSNSGILNV YDRNDLIEKP PKKNESNQEE QNSSEKTKEI RIKKYPTPTR VFEQLTTSIS VVAFSPDGQL LAFGSQ HKK DALRLVHLPS CTVYRNWPTE QTPLGRVTAI AFSSKSDVLA VGNDVGRVRL WEIRG UniProtKB: Uncharacterized protein |
+Macromolecule #15: Putative U3 snoRNP protein
| Macromolecule | Name: Putative U3 snoRNP protein / type: protein_or_peptide / ID: 15 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 115.122008 KDa |
| Sequence | String: MPHPEPDFPL TKRQKLDAAP KASSSKRKKP GSAIFAPYRT IGLISPTGVP FTSIPLGKTT FQITTSVGRA LQTYDLKRGL NLVFVTRPQ TPSDITATHA WKERVYAAFG DPRNGEPQGL WVFQRGKKVA ELPLPSDLDQ PIKQILIFGG WIVACALTRI E VWKAATLE ...String: MPHPEPDFPL TKRQKLDAAP KASSSKRKKP GSAIFAPYRT IGLISPTGVP FTSIPLGKTT FQITTSVGRA LQTYDLKRGL NLVFVTRPQ TPSDITATHA WKERVYAAFG DPRNGEPQGL WVFQRGKKVA ELPLPSDLDQ PIKQILIFGG WIVACALTRI E VWKAATLE HYTTIFPAAS KKGDNELTGG AINMPTFLNK IFVGRKDGWV EIWNVSTGKL IYTLLPPSPD CGAVTCLQPT PA LSLLAIA YSGGPLVIQN VLTDKTVLLL EAGTDDAPVT SISFRTDGLG AGQDGRKDGV MATATSVSGD VTFWDLNKGG RIM GVLRSA HNPPSRHNIV RGGISKIEFL AGQPVIVTSG LDNSLKTWIF DESPFSPVPR ILHQRSGHAA PVRCLHFLPS DFDG AEGGN KWLLSGGKDR SLWGWSLRRD GQSAELSQGA IRKKARKMGL LAGGSHGPTT TLEDLKAPEI TCIASSLNRD GGMGA IPGK QMIWDKGDDK NRISNAELSG NTGWESVVTA HKDDPYARTW FWGRRRAGRW AFKTGDGEPV STVAISSCGT FALVGS TGG SIDMFNLQSG RHRQRFPSRL TPAQLRQLKL QQLRRLDEAN KLAHRSQQKT FAPGTGRHTN AVTGIVVDPL NRHVVSC SL DGKVKFWDFI TGNLVDEIDW APMTKIIGCR YHPGNDLIAF ACDDRSIRVV DIETKNTIRE FWGCRGDIND FCFSPDGR W IVAASQDSII RVWDLPTAHL IDAFRLEQPC TALAFSHTGE YLAGAMEGSL GVQIWTNRTL FRHVPTRQIS ESEIADVAA PTTSGEGGQG LIEAALEAEE EQAEDDGVMA PIIDQLSADM MTLSLVPRSR WQTLLHIDII KARNKPKEPP KAPEKAPFFL PPVGQNGIS SLIPQEDAKA KKEKAAANGA SRITKLDLTR QEQTFTSKLL VGGAKGDYTD FIEHLKALPP AAADLELRSL S IGNGDEAT NELLHFIRAL TSRLVARRDY ELTQAWMTVF LRLHFDLIME NEELLQALGE WREHQARERD RLSELVGYCG GV VSFLRSP RT UniProtKB: Putative U3 snoRNP protein |
+Macromolecule #16: Utp24
| Macromolecule | Name: Utp24 / type: protein_or_peptide / ID: 16 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 22.267188 KDa |
| Sequence | String: MGVAKRTRKF ATVKRIIGKQ DERRKAEAVK KAEEEKRKKE KQAIREVPQM PSSMFFEHNE ALVPPYNVLV DTNFLSRTVG AKLPLLESA MDCLYASVNI IITSCVMAEL EKLGPRYRVA LMIARDERWQ RLTCDHKGTY ADDCIVDRVQ KHRIYIVATN D RDLKRRIR ...String: MGVAKRTRKF ATVKRIIGKQ DERRKAEAVK KAEEEKRKKE KQAIREVPQM PSSMFFEHNE ALVPPYNVLV DTNFLSRTVG AKLPLLESA MDCLYASVNI IITSCVMAEL EKLGPRYRVA LMIARDERWQ RLTCDHKGTY ADDCIVDRVQ KHRIYIVATN D RDLKRRIR KIPGVPIMSV QKGKYAIERL PGAPAS UniProtKB: PIN domain-containing protein |
+Macromolecule #17: Utp30
| Macromolecule | Name: Utp30 / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 43.700527 KDa |
| Sequence | String: MAPSTAVAKK ETDAVIPVDP DQTLKACKAL LAHIKKAAAA PRPDGKQNLL ADEESTVAET PIWLTLTTKK HIHDSHRLQP GKIILPHPL NTSEEISVCL ITADPQRFYK NAVADEFPED LRAKIGRVID ISHLKAKFKA YEAQRKLFSE HDVFLADTRI I NRLPKALG ...String: MAPSTAVAKK ETDAVIPVDP DQTLKACKAL LAHIKKAAAA PRPDGKQNLL ADEESTVAET PIWLTLTTKK HIHDSHRLQP GKIILPHPL NTSEEISVCL ITADPQRFYK NAVADEFPED LRAKIGRVID ISHLKAKFKA YEAQRKLFSE HDVFLADTRI I NRLPKALG KTFYKTTTKR PIPVVLMAQR EKVNGKRVPA PKGKKEKRDP LENANARPIP EIVAEIRKAI GAALVHLSPS TN TAIKVGY ANWEPEKLAA NIETVIRELV ERFVPQKWQN VRNFYVKGPE TAALPIYQTD ELWLDESKVV PDGQEPARAL PGK REKANI GKKRKPLEDA SQPALEETGK DERPKKKAKK TLPESNDDKL DKAIAERKEQ LKKQKAAAKK VAADI UniProtKB: Uncharacterized protein |
+Macromolecule #18: Nop1
| Macromolecule | Name: Nop1 / type: protein_or_peptide / ID: 18 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 32.960844 KDa |
| Sequence | String: MGFERGGRGG GRGGAAARGG RGGARGGRGG PAGRGGPAGR GRGGPAGRGR GGRSGRGGKP KAKGAKAGKK VIVEPHRHKG VFVARGGKE DLLCTANLVP GESVYGEKRI SVETPGSGPD AVATKTEYRI WNPFRSKLAA GILGGLETIY MKPGSKVLYL G AASGTSVS ...String: MGFERGGRGG GRGGAAARGG RGGARGGRGG PAGRGGPAGR GRGGPAGRGR GGRSGRGGKP KAKGAKAGKK VIVEPHRHKG VFVARGGKE DLLCTANLVP GESVYGEKRI SVETPGSGPD AVATKTEYRI WNPFRSKLAA GILGGLETIY MKPGSKVLYL G AASGTSVS HVADIVGPTG AVYAVEFSHR SGRDLINMAT RRTNVIPIVE DARKPMAYRM LVPMVDVIFA DVAQPDQARI VG INARLFL KQGGGLLISI KASCIDSTAP PEQVFASEVQ KLREDKFFPK EQLTLEPYER DHAMVSCVYL QKEFEG UniProtKB: rRNA 2'-O-methyltransferase fibrillarin |
+Macromolecule #19: Putative nucleolar protein
| Macromolecule | Name: Putative nucleolar protein / type: protein_or_peptide / ID: 19 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 57.830699 KDa |
| Sequence | String: MVVVNFLLFE SAVGFSLFEV VHQADTVGLE LPEVKDAMKT LDKFGKMVKL RSFNPWTSAA QGLEAINLIS EGIMPEYLKS ALEMNLPQT SGKKSKVVLG VADKKLAGEI TAAFPGVQCE AADTSEVVAA LLRGIRTHAN KLHKSLQEGD IGRAQLGLGH A YSRAKVKF ...String: MVVVNFLLFE SAVGFSLFEV VHQADTVGLE LPEVKDAMKT LDKFGKMVKL RSFNPWTSAA QGLEAINLIS EGIMPEYLKS ALEMNLPQT SGKKSKVVLG VADKKLAGEI TAAFPGVQCE AADTSEVVAA LLRGIRTHAN KLHKSLQEGD IGRAQLGLGH A YSRAKVKF SVHKNDNHII QGIATLDALD KSINQGAMRV REWYGWHFPE LIRIVSDNIT YAKVVLAIGN KSSLTDESVD DL ANVLNQD QDKALAIIQA AKVSMGQDIS EVDLQMVRDL ASNVTSMADY RRILAESLDK KMSEVAPNLQ VILGTPVAAR LIA HAGSLT NLAKYPASTL QILGAEKALF RALKTKSATP KYGLLYQSSF IGRAGPKVKG RISRYLANKC SIASRIDNFS EKPT RHFGE VLRQQLEQRL EWYAKGTKPM KNSEAMEKAI KAVMADDEET LPVAVDAMDI DSKSPAKEKK DKKEKKEKKE KKEKK KEDK EEKKKDKKRK SLGGEDVEMA DADVGENKKK KKRKSEIAE UniProtKB: Nucleolar protein 56 |
+Macromolecule #20: Nop58
| Macromolecule | Name: Nop58 / type: protein_or_peptide / ID: 20 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 64.292113 KDa |
| Sequence | String: MPLFILTETS AGYALFKAAD KKLLDSDNVS ERLSTLDKII KEIKYKEFAK FDSAAIAVEE ASGILEGKVT PKLASLLNEL KDEKKVTLA VHDTKLSNSI TKLPGINIKP ISGSMTDDLF RAIRQHLYNL IPGMEPSNFD EMNLGLAHSL SRHKLKFSPE K VDVMIVHA ...String: MPLFILTETS AGYALFKAAD KKLLDSDNVS ERLSTLDKII KEIKYKEFAK FDSAAIAVEE ASGILEGKVT PKLASLLNEL KDEKKVTLA VHDTKLSNSI TKLPGINIKP ISGSMTDDLF RAIRQHLYNL IPGMEPSNFD EMNLGLAHSL SRHKLKFSPE K VDVMIVHA VALLDELDKE LNVMAMRVKE WYGWHFPELG KILPDNLSYA RVVLALGLRT NAPNADLSEI LPPEIEAAVK AA ADISMGT EISTEDYENI KLLAVQVVER SEYRRQLAEY LQNRMKAISP NMTELIGALV GARLIAHSGS LVNLAKNPGS TIQ ILGAEK ALFRALKTKH ATPKYGIIYH ASLVGQASGP NKGKIARQLA AKIALSVRTD AFEDFPENAD DETRAAVGIQ ARAK LENNL RLLEGKPLNK GVALGPNGIP VGMPAKWDVK EARKYNIEAD GLATTTSKES SEQPKRPLIE EVPEVEMKDA SASDK KDKK EKKKEKKEVA SSKITEADYE RIAKELGMSL SKFTKKLEKG KIKIKPDGSV EVKGKDGEDA EAETPVKSKS SKRKHE PEE ETPAKEEKHK KKKKKSSKE UniProtKB: Nucleolar protein 58 |
+Macromolecule #21: Snu13
| Macromolecule | Name: Snu13 / type: protein_or_peptide / ID: 21 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 13.831854 KDa |
| Sequence | String: MSNNESAAWP KAEDPALVQE LLDCVQQASH YRQLKKGANE TTKSVNRGTS ELVILAADTQ PLSIVLHIPL ICEEKNVPYV YVPSKVALG RACGVSRAVI AVSLTSNEAS DLNSKIRALR DKVERLAM UniProtKB: H/ACA ribonucleoprotein complex subunit 2 |
+Macromolecule #22: Rrp9
| Macromolecule | Name: Rrp9 / type: protein_or_peptide / ID: 22 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 69.610438 KDa |
| Sequence | String: MSSFFTAPAS EKKRKRAATA DAPKKRLATT KSSSKSGSKA PTKAAAPASK KKAIERDESI SGSDLDSDLS GDDEFIERRS SDAGSGDES EKEGETAAEK RLRLAQRYLE KTRKEVEQLD EYAFDAEEID RDLLAERLKE DVAEAKGKVY RRLASELAFD K ATYTQFRW ...String: MSSFFTAPAS EKKRKRAATA DAPKKRLATT KSSSKSGSKA PTKAAAPASK KKAIERDESI SGSDLDSDLS GDDEFIERRS SDAGSGDES EKEGETAAEK RLRLAQRYLE KTRKEVEQLD EYAFDAEEID RDLLAERLKE DVAEAKGKVY RRLASELAFD K ATYTQFRW NSGTVTSVAV CPPYAYTTTK DGYLTKWKLQ DLPKNQWPQT TKKKPKKPPA PPKKRPERIC FAKADARKAN DK TYQGHLK APLVVKASQD GKFVVTGGAD KRLVVYNAAD LKPIKAFTQH RDAVTGLAFR RGTNQLYSCS KDRTVKVWSL DEL AYVETL FGHQDEILDI DALGQERCVS VGARDRTARY WKVPEESQLV FRGGGEGGSS NTKKHKLPPG MDPASAAHEG SMDR VAMID DDMFVTGSDN GDLALWSIQR KKPLHVIARA HGLEPPIKLE DYSADEIPDP SIIPAPQPRG ITALRTLPYS DLIFS GSWD GCIRVWRLSE DKRKLEAVGI LGVGSETCEN STNISNGATS NGESSSTSSS TTLAAQSSSS SSPPPSQPKD LVRGIV NDI ALFERGERGR DGLCVVVVTG KEMRFGRWKY MKEGRCGAVI FEVPKVEKKN KRKNEDKKEE VNGVYKE UniProtKB: Ribosomal RNA-processing protein |
+Macromolecule #23: RNA 3'-terminal phosphate cyclase-like protein
| Macromolecule | Name: RNA 3'-terminal phosphate cyclase-like protein / type: protein_or_peptide / ID: 23 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 43.399766 KDa |
| Sequence | String: MAEPKPEFLR FTGHRAFTQR LVLATLYGRP IHISKIRSSS ATNPGLAPHE ISFLRLLESV TNGSIIDVSY SGTTITYQPG LITGTVPGM NASLSSDAIE HVIPATNTRG ITYFLIPLAL LAPFSKAHLN VRFTGPGVIT SATHGARDLS IDTFRTAVLP L YGLFGIPP ...String: MAEPKPEFLR FTGHRAFTQR LVLATLYGRP IHISKIRSSS ATNPGLAPHE ISFLRLLESV TNGSIIDVSY SGTTITYQPG LITGTVPGM NASLSSDAIE HVIPATNTRG ITYFLIPLAL LAPFSKAHLN VRFTGPGVIT SATHGARDLS IDTFRTAVLP L YGLFGIPP ARIELRVLQR SCAGPGGKGG GGIVEMRFAS QVRLPKTLHL NRRPGKVRRI RGVAYCTGVA ASHNNRMITA AR GVLNQLV SDVHIAAQYD PAPLVAEKGT TQKKKTGIGF GLSLVAETSA EGVIYAADEV APPEGGVVPE DIGEKCAYQL LDV IAQGGC VMAASAPTVL TLMAMGSEDV GRLRLGRRVV SPELLELARD LKAFGAASWG IRDAGDDEDD AEGELGDLIV SVKG TGVGN VGRKVA UniProtKB: RNA 3'-terminal phosphate cyclase-like protein |
+Macromolecule #24: Bms1
| Macromolecule | Name: Bms1 / type: protein_or_peptide / ID: 24 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 134.17225 KDa |
| Sequence | String: MDSQQHKPHR PSKTKEKKKK QNSGGTNPKA FAVANPGKLA RQAARSHDIK EKRLHVPLVD RLPDEPPPRL VVIVGPPGVG KTTLLKSLV RRYTKETMSD PVGPITVVTS KKQRLTFIEC PNELEAMIDM AKVADIVLLM IDGNYGFEME TMEFLNILAN T GMPGNVFG ...String: MDSQQHKPHR PSKTKEKKKK QNSGGTNPKA FAVANPGKLA RQAARSHDIK EKRLHVPLVD RLPDEPPPRL VVIVGPPGVG KTTLLKSLV RRYTKETMSD PVGPITVVTS KKQRLTFIEC PNELEAMIDM AKVADIVLLM IDGNYGFEME TMEFLNILAN T GMPGNVFG ILTHLDLFKK PSALKDAKKR LKHRLWTELY QGAHLFYLSG VLNGRYPDRE IHNLSRFLSV MKNPRPLVWR NT HPYTIID NYRDITHPTK IEEDPLCDRT IELSGYLRGT NFAAQGQRVH IAGVGDFTIS KIEELPDPCP TPAMEKAMAN LTG KKPRRR LDEKDKKLWA PMADRSGMKI SGDHIVITRE KGFTFDKDAN VERGEGEQLI VDLQGEKKLL GQTDKGVKLF AGGE QLTQI PEEDTNDTGR KTHRKARFLE DDRQNDENGV PEDEGFVSGE ESKGSDGSDI EEEFDEKRLG KMFRGDDEET QDEDV VFAD TDSELGSISG DEGVDSEESG SDEEFDSEEE EAVRWKENMM ERARALHGKR KPWRAIDLAR LMYDTTLTPA QALRRW RGE DNEKEEEEEE DIEKDEDTFF HKAKDEDDLE EDRMIPHFDY EELKTKWSNP ENIDALRRTR FSTGRPKGDG EGDSEGE GD GDDDDDFNGF DEDDEDEGDG AFEDLETGEK HGPSKEKKEE EEKPAMSLEE ERERNARRKE ELRARFEEED REGLLNDK A IARREGGLDE EFGEDQWYEM QKAMLQKQLD INKAEYAELD EHQRRQVEGY RAGKYARLVI EGVPAEFCKN FQPRMPILV GGLSATEDRF GFVQVRIKRH RWHKKILKTG DPLIFSLGWR RFQTLPIYSI WDNRTRNRML KYTPEHMHCF GTFWGPLIAP NTSFCCFQS FSASNPGFRI AATGTVLSVD ESTEIVKKLK LVGTPWKIFK NTAFIKDMFN SSLEIAKFEG AAIRTVSGIR G QIKRALSK PEGYFRATFE DKILLSDIVI LKAWYPVKPK QFYNPATNLI GWQSMRLTGE IRRAENIPTP QNPNSTYRKI ER PERHFNP LRVPKNLAAE LPFKSQIVQT KPQKKETYMQ KRAVVVGREE RKLRDLMQKL TTIRKEKIAK RKAKKEAQRE KLK KELAEI EERRREKQKK EKKEFWEREG KKRKASEEWG GGGKRRK UniProtKB: Bms1-type G domain-containing protein |
+Macromolecule #25: Imp3
| Macromolecule | Name: Imp3 / type: protein_or_peptide / ID: 25 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 21.802396 KDa |
| Sequence | String: MVRKLKYHEQ KLLKKHDFIN YKSDNNHRDH DVIRRYMIQK PEDYHKYNRL CGSLRQFAHR LSLLPPDNEV RRKHETLLLD KLYDMGILS TKAKLSAVEH NVTVSAFARR RLPVVMTRLR MAETVQAATK LIEQGHVRVG VEEVRDPAFL VTRNMEDFVT W TVGSKIKQ NIMKYRDKLD DFELL UniProtKB: U3 small nucleolar ribonucleoprotein protein IMP3 |
+Macromolecule #26: Putative U3 small nucleolar ribonucleoprotein
| Macromolecule | Name: Putative U3 small nucleolar ribonucleoprotein / type: protein_or_peptide / ID: 26 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 34.216266 KDa |
| Sequence | String: MLRKQARQRR DYLYRRALLL RDAEIAEKRA KLRAALASGK PLDPKIANDK ELRKDFDYDV SRDIAKEQGE IDIDDEYSEL SGIVDPRVL VTTSRDPSSR LMAFSKEIRL MFPTAIRLNR GNLILPDLVM SAQRERLSDI ILLHEHRGTP TAITISHFPH G PTLMASLH ...String: MLRKQARQRR DYLYRRALLL RDAEIAEKRA KLRAALASGK PLDPKIANDK ELRKDFDYDV SRDIAKEQGE IDIDDEYSEL SGIVDPRVL VTTSRDPSSR LMAFSKEIRL MFPTAIRLNR GNLILPDLVM SAQRERLSDI ILLHEHRGTP TAITISHFPH G PTLMASLH NVVLRADIPK SIKGTVSESY PHLIFEGFRT PLGQRVVKIL KHLFPPRDPT NNAKSGNRVI TFVNQDDCIE VR HHVYVRT NYNSVELSEV GPRFTMRPFS ITMGTLENKD ADVEWHLSQY TRTGRKKNYF UniProtKB: U3 small nucleolar ribonucleoprotein protein IMP4 |
+Macromolecule #27: Putative U3 small nucleolar ribonucleoprotein protein
| Macromolecule | Name: Putative U3 small nucleolar ribonucleoprotein protein / type: protein_or_peptide / ID: 27 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 86.082141 KDa |
| Sequence | String: MPGAPSTTSS FTSTSHTLSA LPSMPQSLSA SAADGSGSGS DGAAAAAITA FLDSTAPENR HIFLRPTPQL PAGSLALLKA ALDPLAAQI ADHQAAGIAR LRESGALSSK KRKRDGSEKE NKPAALKIRK VHVDGFETQQ VWQQARKIIT SALGEAQAVL E ELKVNGEV ...String: MPGAPSTTSS FTSTSHTLSA LPSMPQSLSA SAADGSGSGS DGAAAAAITA FLDSTAPENR HIFLRPTPQL PAGSLALLKA ALDPLAAQI ADHQAAGIAR LRESGALSSK KRKRDGSEKE NKPAALKIRK VHVDGFETQQ VWQQARKIIT SALGEAQAVL E ELKVNGEV EEEEGEDKVI EFGEDGFEVG SSDEEESEEE GNEEADTEDS DGEGASLGDE NAMFDLEAEE DSGSEEDKSD VG EEVDGEV DGEKHSDLDG EEEGEEGEED EEDEEDDDES AEDLVEDPHG LNDGFFSIDE FNKQTQMWED QDMRAEPTAE LDD DSEDID WHADPFAVKP SKRGKKDDGD MDLDDEEDES DDEAPPVGKK ALEKMLDKDE DDEGGNLEDD LADGMGMDLT ANDI YYKDF FAPPRKKKKP GSSKKKRELE LETKRPDDAD VERAEQDVRR DLFDDLSEHE DSEDALSDAS AGDPKSRKSA HERRQ AKIA EQIRKLEAEL VAKRAWTLAG EATAADRPVN SLLGEDMEFD HVGKPVPVVT EEVSESIEEL IKRRILAGEF DEVLRR RPD MFGNPHGVRR GLVDVEDTKA KQSLAEIYEE EAVKKANPDA YVSAADEKLR RDEEEIKRMW KEISAKLDAL SSWHYKP KP PAPTLTVVSD VATVAMEDAQ PATAQGVAGG ETSMIAPQEV YAPSKDTAEK GEVVTKAGIP IAKQEMSREE KLRRRRRE K ERIRKAGGLD GGKPVSEKEK EKKETVAQLK KSGVKVINRK GEVVGLDGKK VGEKKVQSSG AYKL UniProtKB: Putative U3 small nucleolar ribonucleoprotein protein |
+Macromolecule #28: Sof1
| Macromolecule | Name: Sof1 / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 50.869828 KDa |
| Sequence | String: MKIKALTRSI TAQQAPGSDV QRAPRNLAPE LHPFERAREY QRALNAVKLE RMFAKPFLGQ LGNGHVQGVY SMCKDKNSLN CIASGSGDG VVKVWDLTTR DEETWRVAAH NNIVKGLTFT NDKKLLSCAT DGIKLWDPYA SPSNTTPIAT WQEGGPYTSL S FHRSANTF ...String: MKIKALTRSI TAQQAPGSDV QRAPRNLAPE LHPFERAREY QRALNAVKLE RMFAKPFLGQ LGNGHVQGVY SMCKDKNSLN CIASGSGDG VVKVWDLTTR DEETWRVAAH NNIVKGLTFT NDKKLLSCAT DGIKLWDPYA SPSNTTPIAT WQEGGPYTSL S FHRSANTF AASSGQGCIR IWDLEHSTAG QAIQWPSFVD TITDVCFNQV ETSVIGSVAT DRSIILFDLR TNMPVIKTVL HF ACNRIVF NPMEAMNLAV ASEDHNIYIF DARNFDKALN IQKGHVAAVM DVEFSPTGEE LVSGSYDRTI RLWRRDAGHS RDV YHTKRM QRVFRTMWTM DSKYILTGSD DGNVRLWRAN ASERSGVKAT RQRQALEYNN ALLDRYGHLP EIRRIRRHRH LPKV VKKAT EIKREELAAI KRREENERKH SNKKYEKRKS EREKAVLVKQ Q UniProtKB: DDB1- and CUL4-associated factor 13 |
+Macromolecule #29: Emg1
| Macromolecule | Name: Emg1 / type: protein_or_peptide / ID: 29 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 27.936164 KDa |
| Sequence | String: MQSQTAGTQS LPPPALPQLV AEQHVPIPPN DKDTKRLIVV LSNASLETYK ASHGTNRNGV REEKYTLLNS DEHIGIMRKM NRDISDARP DITHQCLLTL LDSPINKAGK LQIYIQTAKG VLIEVSPTVR IPRTFKRFAG LMVQLLHRLS IKGTNTNEKL L KVIQNPIT ...String: MQSQTAGTQS LPPPALPQLV AEQHVPIPPN DKDTKRLIVV LSNASLETYK ASHGTNRNGV REEKYTLLNS DEHIGIMRKM NRDISDARP DITHQCLLTL LDSPINKAGK LQIYIQTAKG VLIEVSPTVR IPRTFKRFAG LMVQLLHRLS IKGTNTNEKL L KVIQNPIT DHLPPNCRKV TLSFDAPLVR VRDYVDTLGP NESICVFVGA MAKGPDNFAD AYVDEKISIS NYSLSASVAC SK FCHACED AWDII UniProtKB: Nucleolar essential protein 1 |
+Macromolecule #30: KRR1 small subunit processome component
| Macromolecule | Name: KRR1 small subunit processome component / type: protein_or_peptide / ID: 30 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 37.500777 KDa |
| Sequence | String: MPSTHKKDKP WDTDDIDKWK IEPFLPEHSS GPFLEESSFM TLFPKYRERY LKDCWPLVTK ALEKHGIAAT LDIVEGSMTV KTTRKTYDP AAILKARDLI KLLARSVPAP QALKILEDGM ACDIIKIRSM VRNKERFVKR RQRLLGQNGT TLKALELLTQ T YILVHGNT ...String: MPSTHKKDKP WDTDDIDKWK IEPFLPEHSS GPFLEESSFM TLFPKYRERY LKDCWPLVTK ALEKHGIAAT LDIVEGSMTV KTTRKTYDP AAILKARDLI KLLARSVPAP QALKILEDGM ACDIIKIRSM VRNKERFVKR RQRLLGQNGT TLKALELLTQ T YILVHGNT VSVMGGYKGL KEVRRVVEDT MNNIHPIYLI KELMIKRELA KDPALAHEDW SRYLPQFKKR TLSKRRKPFK IN DKSKKPY TPFPPAPEKS KIDLQIESGE YFLSKEAKQR AAEAERAEKA RQKKEEKKRE REKEFVPPEE DGGKKKKRKV KHG EE UniProtKB: KRR1 small subunit processome component |
+Macromolecule #31: Pre-rRNA-processing protein PNO1
| Macromolecule | Name: Pre-rRNA-processing protein PNO1 / type: protein_or_peptide / ID: 31 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 28.715047 KDa |
| Sequence | String: MPAPTALKQP PPAPEQQAAP AITNENEDEL LIDIQQAAAT LTDPNAAEPP EETMENEMAV DEEGRPRFAP GKNIDPIRRI ETRKIPIPP HRMSALKANW TKYPPLVDHC KLQVRMNIKE KRVELRSSKY TVSNEALQMG ADFVSAFAMG FDIDDAIALL R LDSLYIQS ...String: MPAPTALKQP PPAPEQQAAP AITNENEDEL LIDIQQAAAT LTDPNAAEPP EETMENEMAV DEEGRPRFAP GKNIDPIRRI ETRKIPIPP HRMSALKANW TKYPPLVDHC KLQVRMNIKE KRVELRSSKY TVSNEALQMG ADFVSAFAMG FDIDDAIALL R LDSLYIQS FDIKDVRQTL GPDALSRAIG RIAGKDGKTK FAIENATKTR IVLAGSKVHI LGAFENIGMA RESIVSLVLG AQ PGKVYNN LRIIASRMKE RW UniProtKB: Pre-rRNA-processing protein PNO1 |
+Macromolecule #32: RNA cytidine acetyltransferase
| Macromolecule | Name: RNA cytidine acetyltransferase / type: protein_or_peptide / ID: 32 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 119.795125 KDa |
| Sequence | String: MTVQKTVDSR IPTLIRNGLQ TKKRSFFVVV GDHAKEAIVH LYYIMSSMDV RQNKSVLWAY KKELLGFTSH RKKREAKIKK EIKRGIREP NQADPFELFI SLNDIRYCYY KETDKILGNT YGMCILQDFE AITPNILART IETVEGGGLV VLLLKGMTSL K QLYTMTMD ...String: MTVQKTVDSR IPTLIRNGLQ TKKRSFFVVV GDHAKEAIVH LYYIMSSMDV RQNKSVLWAY KKELLGFTSH RKKREAKIKK EIKRGIREP NQADPFELFI SLNDIRYCYY KETDKILGNT YGMCILQDFE AITPNILART IETVEGGGLV VLLLKGMTSL K QLYTMTMD VHARYRTEAH DDVIARFNER FLLSLGSCES CLVIDDELNV LPISGGKGVK PLPPPDEDEE LSPAAKELKK IK DELEDTQ PIGSLIKLAR TVDQAKALLT FVDAIAEKTL RNTVTLTAAR GRGKSAAMGV AIAAAVAYGY SNIFITSPSP ENL KTLFEF VFKGFDALDY KDHADYTIIQ STNPEFNKAI VRVNIHRNHR QTIQYIRPQD AHVLGQAELV VIDEAAAIPL PLVK KLMGP YLVFMASTIS GYEGTGRSLS LKLIKQLREQ SRAGANPNGG NAVEVDRSTL KATKETTSVG GRSLKEITLS EPIRY AQGD NVEKWLNTLL CLDATLPRSK ISTTGCPDPS QCELLHVNRD TLFSFHPVSE KFLQQMVALY VASHYKNSPN DLQLMS DAP AHELFVLTGP IQEGRLPEPL CVIQVSLEGK ISKQSILKSL SRGQQPAGDL IPWLVSQQFQ DDEFASLSGA RIVRIAT NP DYMSMGYGSK ALQLLVDYYE GKFADLSEDA AAEVPRSIPR VTDAELSKGS LFDDIKVRDM HELPPLFSKL SERRPEKL D YVGVSYGLTQ QLHKFWKRAQ FVPVYLRQTA NDLTGEHTCV MIRPLQDGND PSWLGAFAAD FHKRFLSLLS YKFREFPSI LALTIEESAN AGAMLDPSNA PTELTKAELD QLFTPFDHKR LESYANGLLD YHVVLDLMPT IAQLYFTGRL REAVKLSGLQ QAILLALGL QRKDIDTLAT ELNLPGSQVL AIFMKIMRKV TQHFGALVSG AIAAELPDPN KTVGVSKENA MGIHDDEVVG L KFEALEQR LEDELDEGGD EALRELRKKQ RELIDSLPLD QYEIDGDDDA WKEAEKRVAS AAKSGKKVDG TLVSVPSAKA AK RKAEEMA ALRDELEKME KGKERGSKKA KKEKRR UniProtKB: RNA cytidine acetyltransferase |
+Macromolecule #33: Fcf2
| Macromolecule | Name: Fcf2 / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 22.569129 KDa |
| Sequence | String: MATTLGLPDE EIDRLLAEAE ARLAGSGDAD AGAIALAKPA ASKPLTVAAP AAPKAGEQTV PQVKKAEELS VRVPQLPQKK KGPPDTLSD WYNIPRTNLT PELKRDLQLL RMRDVVAMGK QFFKKDNRKD FVPEYCQVGT IIAGATDGVS GRLTRKERKR T IVEEVLSS ...String: MATTLGLPDE EIDRLLAEAE ARLAGSGDAD AGAIALAKPA ASKPLTVAAP AAPKAGEQTV PQVKKAEELS VRVPQLPQKK KGPPDTLSD WYNIPRTNLT PELKRDLQLL RMRDVVAMGK QFFKKDNRKD FVPEYCQVGT IIAGATDGVS GRLTRKERKR T IVEEVLSS DSVSKYKRKY HEIQEHKKSG RKGYYKKLMA ARKRK UniProtKB: Fcf2 pre-rRNA processing C-terminal domain-containing protein |
+Macromolecule #34: 40S ribosomal protein S1
| Macromolecule | Name: 40S ribosomal protein S1 / type: protein_or_peptide / ID: 34 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 29.245158 KDa |
| Sequence | String: MAVGKNKRLS KGKKGLKKKV QDPFTRKDWY NIKAPAPFAV RDVGKTLVNR TTGLKNANDA LKGRIFEVCL ADLQKDEDHA FRKIKLRVD EVQGKNCLTN FHGLDFTTDK LRSLVRKWQT LIEANVTVTT TDHYLLRLFA IAFTKRRPNQ IKKTTYAQSS Q IRAIRRKM ...String: MAVGKNKRLS KGKKGLKKKV QDPFTRKDWY NIKAPAPFAV RDVGKTLVNR TTGLKNANDA LKGRIFEVCL ADLQKDEDHA FRKIKLRVD EVQGKNCLTN FHGLDFTTDK LRSLVRKWQT LIEANVTVTT TDHYLLRLFA IAFTKRRPNQ IKKTTYAQSS Q IRAIRRKM VEIIQREAAS CTLHQLVSKL IPEVIGREIE KATQGIYPLQ NVHIRKVKLL KQPKFDLGAL MALHGESSEE AG QKVEREF REQVLESV UniProtKB: Small ribosomal subunit protein eS1 |
+Macromolecule #35: 40S ribosomal protein S4
| Macromolecule | Name: 40S ribosomal protein S4 / type: protein_or_peptide / ID: 35 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 29.800764 KDa |
| Sequence | String: MARGPKKHQK RLSAPSHWLL DKLSGAYAPR PSTGPHKLRD CMPLIVFVRN RLKYALNYRE TKAIMMQRLV KVDGKVRTDI TYPAGFMDV ITIEKTGENF RLIYDTKGRF TVHRITDEEA KYKLGKVKRV QLGRGGVPFL VTHDARTIRY PDPLIKVNDT V KIDLETGK ...String: MARGPKKHQK RLSAPSHWLL DKLSGAYAPR PSTGPHKLRD CMPLIVFVRN RLKYALNYRE TKAIMMQRLV KVDGKVRTDI TYPAGFMDV ITIEKTGENF RLIYDTKGRF TVHRITDEEA KYKLGKVKRV QLGRGGVPFL VTHDARTIRY PDPLIKVNDT V KIDLETGK ITDFIKFDTG ALAMITGGRN MGRVGVITHR ERHDGGFGIV HLKDALDNTF ATRESNVFVI GSEKPWISLP KG KGVKLTI AEERDQRRAR ALAAAGH UniProtKB: 40S ribosomal protein S4 |
+Macromolecule #36: 40S ribosomal protein s5-like protein
| Macromolecule | Name: 40S ribosomal protein s5-like protein / type: protein_or_peptide / ID: 36 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 23.679225 KDa |
| Sequence | String: MSEGEVEVAQ PQYDVLPKEV LAEVGSVKLF NRWSYDDIEI RDISLTDYIQ IRAPVYIPHS AGRYAVKRFR KANCPIIERL TNSLMMHGR NNGKKLMAVR IVAHAFEIIH LMTDQNPIQV AVDAIVNCGP REDSTRIGSA GTVRRQAVDV SPLRRVNQAI A LLTTGARE ...String: MSEGEVEVAQ PQYDVLPKEV LAEVGSVKLF NRWSYDDIEI RDISLTDYIQ IRAPVYIPHS AGRYAVKRFR KANCPIIERL TNSLMMHGR NNGKKLMAVR IVAHAFEIIH LMTDQNPIQV AVDAIVNCGP REDSTRIGSA GTVRRQAVDV SPLRRVNQAI A LLTTGARE AAFRNVKTIA ECLAEELINA AKGSSNSYAI KKKDELERVA KSNR UniProtKB: 40S ribosomal protein s5-like protein |
+Macromolecule #37: 40S ribosomal protein S6
| Macromolecule | Name: 40S ribosomal protein S6 / type: protein_or_peptide / ID: 37 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 27.490139 KDa |
| Sequence | String: MKLNISHPAN GSQKLIEVED ERKLRHFYDK RMGAEVAGDP LGPEWKGYIL RITGGNDKQG FPMKQGVIAP NRVRLLLSEG HSCYRPRRD GERKRKSVRG CIVGPDLSVL ALSIVKQGEQ DIPGLTDVVH PKRLGPKRAT KIRRFFSLSK DDDVRKYVIR R EVQPKGEG ...String: MKLNISHPAN GSQKLIEVED ERKLRHFYDK RMGAEVAGDP LGPEWKGYIL RITGGNDKQG FPMKQGVIAP NRVRLLLSEG HSCYRPRRD GERKRKSVRG CIVGPDLSVL ALSIVKQGEQ DIPGLTDVVH PKRLGPKRAT KIRRFFSLSK DDDVRKYVIR R EVQPKGEG KKPYTKAPRI QRLVTPQRLQ HKRHRIALKR RQQEKVKEEA AEYAQILAKR VAEAKAQKAD LRKRRASSLH K UniProtKB: 40S ribosomal protein S6 |
+Macromolecule #38: 40S ribosomal protein S7-like protein
| Macromolecule | Name: 40S ribosomal protein S7-like protein / type: protein_or_peptide / ID: 38 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 23.08465 KDa |
| Sequence | String: MSAPSLNKIA ANSPSRQNPS DLERAIAGAL YDLETNTADL KAALRPLQFV SAREIEVGHG KKAIVIFVPV PALQGFHRVQ QRLTRELEK KFSDRHVLIL AARRILPKPK RSARSRNTLK QKRPRSRTLT AVHDAILTDL VYPVEIVGKR LRTKEDGSKV L KVILDEKE ...String: MSAPSLNKIA ANSPSRQNPS DLERAIAGAL YDLETNTADL KAALRPLQFV SAREIEVGHG KKAIVIFVPV PALQGFHRVQ QRLTRELEK KFSDRHVLIL AARRILPKPK RSARSRNTLK QKRPRSRTLT AVHDAILTDL VYPVEIVGKR LRTKEDGSKV L KVILDEKE RGGVDYRLDT YSEVYRRLTG RNVTFEFPQT TITDY UniProtKB: 40S ribosomal protein S7 |
+Macromolecule #39: 40S ribosomal protein S8
| Macromolecule | Name: 40S ribosomal protein S8 / type: protein_or_peptide / ID: 39 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 23.102416 KDa |
| Sequence | String: MGISRDSRHK RSHTGAKRAF YRKKRAFELG RQPANTRIGP KRIHIVRTRG GNHKYRALRL DSGNFAWGSE GCTRKTRIIG VVYHPSNNE LVRTNTLTKS AVVQIDAAPF RQWYEAHYGQ PLGRRRQQKQ GQVVEEVKKS KSVEKKQAAR FAAHGKVDPA L EKQFEAGR ...String: MGISRDSRHK RSHTGAKRAF YRKKRAFELG RQPANTRIGP KRIHIVRTRG GNHKYRALRL DSGNFAWGSE GCTRKTRIIG VVYHPSNNE LVRTNTLTKS AVVQIDAAPF RQWYEAHYGQ PLGRRRQQKQ GQVVEEVKKS KSVEKKQAAR FAAHGKVDPA L EKQFEAGR LYAIISSRPG QSGRADGYIL EGEELAFYQR KLHK UniProtKB: 40S ribosomal protein S8 |
+Macromolecule #40: 40S ribosomal protein s9-like protein
| Macromolecule | Name: 40S ribosomal protein s9-like protein / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 22.029836 KDa |
| Sequence | String: MAPRKYSKTY KVPRRPYEAA RLDSELKLVG EYGLRNKREV WRVLLTLSKI RRAARILLTL DEKDPKRLFE GNALIRRLVR IGVLDESRM KLDYVLALKA EDFLERRLQT LVYKLGLAKS IHHARVLIRQ RHIRVGKQIV NVPSFMVRLD SQKHIDFALT S PFGGGRPG RVRRKKAKAA EGGEGGEEEE EE UniProtKB: 40S ribosomal protein s9-like protein |
+Macromolecule #41: 40S ribosomal protein S13-like protein
| Macromolecule | Name: 40S ribosomal protein S13-like protein / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 16.912891 KDa |
| Sequence | String: MGRLHSKGKG ISASAIPYSR NPPAWLKTTP EQVVEQICKL ARKGATPSQI GVILRDSHGI AQVKVVTGNK ILRILKSNGL APDIPEDLY FLIKKAVAVR KHLERNRKDK DSKFRLILIE SRIHRLARYY KTVGVLPPTW KYESSTASTL VA UniProtKB: 40S ribosomal protein S13-like protein |
+Macromolecule #42: 40S ribosomal protein S14-like protein
| Macromolecule | Name: 40S ribosomal protein S14-like protein / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 16.071464 KDa |
| Sequence | String: MPPKKTTRPA QENISLGPQV REGELVFGVA RIFASFNDTF VHVTDLSGRE TICRVTGGMK VKADRDESSP YAAMLAAQDV AARCKELGI TALHIKIRAT GGNGTKTPGP GAQSALRALA RSGMKIGRIE DVTPTPSDST RRKGGRRGRR L UniProtKB: 40S ribosomal protein S14-like protein |
+Macromolecule #43: 40S ribosomal protein S16-like protein
| Macromolecule | Name: 40S ribosomal protein S16-like protein / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 15.962771 KDa |
| Sequence | String: MASVQAVQVF GKKKNATAVA RCVQGKGLIK VNGKPLKLFA PEILRAKLYE PILILGTDKF ADVDIRIRVA GGGHTSQVYA VRQAIAKSI VAYYAKYVDE HSKNLLKQEL IQFDRSLLVA DPRRCEPKKF GGRGARARFQ KSYR UniProtKB: 40S ribosomal protein S16-like protein |
+Macromolecule #44: 40S ribosomal protein S11-like protein
| Macromolecule | Name: 40S ribosomal protein S11-like protein / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 18.698182 KDa |
| Sequence | String: MATELTVQSE RAFLKQPHIF LNSKVKVKST RPGKGGRRWY KDVGLGFKTP KTAIEGHYID KKCPFTGMVS IRGRILTGRV VSTKMHRTI IIRREYLHYI PKYNRYEKRH KNLAAHVSPA FRVEEGDMVV VGQCRPLSKT VRFNVLRVLP RTGKSVKKFQ K F UniProtKB: 40S ribosomal protein S11-like protein |
+Macromolecule #45: 40S ribosomal protein S22-like protein
| Macromolecule | Name: 40S ribosomal protein S22-like protein / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 14.905471 KDa |
| Sequence | String: MVRTSVLHDA LNSINNAEKM GKRQVMIRPS SKVIVKFLQV MQRHGYIGEF EEVDNHRSGK IVVQLNGRLN KCGVISPRYN VRLAELEKW VTKLLPARQF GYVILTTSAG IMDHEEARRK HVAGKIIGFF Y UniProtKB: 40S ribosomal protein S22-like protein |
+Macromolecule #46: 40S ribosomal protein s23-like protein
| Macromolecule | Name: 40S ribosomal protein s23-like protein / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 15.934653 KDa |
| Sequence | String: MSGGKPRGLN AARKLRNNRR EQRWADLQYK KRALGTAYKS SPFGGSSHAK GIVLEKVGVE AKQPNSAIRK CVRVQLIKNG KKVTAFVPN DGCLNFVDEN DEVLLAGFGR KGKAKGDIPG VRFKVVKVSG VGLLALWKEK KEKPRS UniProtKB: 40S ribosomal protein s23-like protein |
+Macromolecule #47: 40S ribosomal protein S24
| Macromolecule | Name: 40S ribosomal protein S24 / type: protein_or_peptide / ID: 47 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 15.535286 KDa |
| Sequence | String: MADTDSPVTL RTRKFIRNPL LSRKQMVVDI LHPGRPNISK DELREKLATM YKAQKDQVSV FGLRTQYGGG KTTGFALIYD SPEALKKFE PRYRLVRAGL AAKVEKASRQ QRKQRKNRLK TLRGTAKVKG AKAKKEK UniProtKB: Small ribosomal subunit protein eS24 |
+Macromolecule #48: 40S ribosomal protein S28-like protein
| Macromolecule | Name: 40S ribosomal protein S28-like protein / type: protein_or_peptide / ID: 48 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 7.74198 KDa |
| Sequence | String: MDSSKAPVKF VKVTRVLGRT GSRGGVTQVR VEFMDDTTRS IIRNVKGPVR ENDILVLLES EREARRLR UniProtKB: 40S ribosomal protein S28-like protein |
+Macromolecule #49: Faf1
| Macromolecule | Name: Faf1 / type: protein_or_peptide / ID: 49 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 34.576586 KDa |
| Sequence | String: MPGLLGKRKS RAEEDPEAVA KAQELLRKHF EAQFKPIDLA PLPRRAIESE DEEDESSEEG SDVNSGEGDE WDGISGDEDG TESEGDESD DEPHVVQVVD YSNDSSAADG KMSKQELKVY LSSRPPDPTR KSSSSKPKPS KKSTDDSFPE DSAELLANDL A LQRLIAES ...String: MPGLLGKRKS RAEEDPEAVA KAQELLRKHF EAQFKPIDLA PLPRRAIESE DEEDESSEEG SDVNSGEGDE WDGISGDEDG TESEGDESD DEPHVVQVVD YSNDSSAADG KMSKQELKVY LSSRPPDPTR KSSSSKPKPS KKSTDDSFPE DSAELLANDL A LQRLIAES HILSEAGANP SHWQSSHAAT TGTNTRAFAT GRIAKKTTDM RIQALGAKES ILTQQKMPMN MRKGIVKHQE EK EKKRRQE ARENGIVLER EVKKKKTVRK RRERPVDLPA VGRMRGAELR ISAKEAAAIA REVRGPQGRG KRRR UniProtKB: Uncharacterized protein |
+Macromolecule #50: 35S rRNA
| Macromolecule | Name: 35S rRNA / type: rna / ID: 50 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 827.801812 KDa |
| Sequence | String: GGGGGGAUGU CCGGGUCGCC CUGGGCACCU CUCCGCGGCA GCCUUCGCGG GCAAGCCGUA GUCCCGGUUC AUACGUCGCC GCAAUCGCG AGGGGACCAA CGGGUAACCG UUCCUCCAAG CGGUCGCCGG CGAGCACCAC CCCAGCCCCG CCGCCGCCCC C AGCGCACC ...String: GGGGGGAUGU CCGGGUCGCC CUGGGCACCU CUCCGCGGCA GCCUUCGCGG GCAAGCCGUA GUCCCGGUUC AUACGUCGCC GCAAUCGCG AGGGGACCAA CGGGUAACCG UUCCUCCAAG CGGUCGCCGG CGAGCACCAC CCCAGCCCCG CCGCCGCCCC C AGCGCACC GCGCGCAGGG GGGCCGGCGC GGCAUCUGGG AAGACUCCUA AGGCGCAAGG GAAUACCCCG UGUGCGCCGG GG GUUGGGC UCUGUGGGUG CUUGGCCGCU GGCCAGACCU ACUUCAAAGC CACCGGGGGU ACCUACCGGU UGGGGAAGCC CUA GCGGCU GAACCUCUCG GUCGGGCCUC CUCUUGGGGC GCCUAGGCGG UGUCUUCGGG UCGCGUGCAG CCUCCCGUCC UCGC CUGCC GACAGCGCUC UGGGGAAUUC AGGAGGGGAA AGCAGAUGUG CCCCGCGGCG AGACUGGCGC ACCGCCGGAC GAGCU GCUG GGGCACGCGG GUACGCUAGC CGCUAGCGCC CGCCAGCCUG UAACUAGAGU GAACUCGGCC GCCCUGAAAC ACGGGC GGU UGGCCUGCUU AGGCGCACGA UAGUUACCUG GUUGAUUCUG CCAGUAGUCA UAUGCUUGUC UCAAAGAUUA AGCCAUG CA UGUCUAAGUA UAAGCAAUUA UACAGCGAAA CUGCGAAUGG CUCAUUAAAU CAGUUAUCGU UUAUUUGAUA GUACCUUA C UACAUGGAUA ACCGUGGUAA UUCUAGAGCU AAUACAUGCU GAAAAUCCCG ACUUCGGAAG GGAUGUAUUU AUUAGAUUA AAAACCAAUG CCCUUCGGGG CUUUCUGGUG AUUCAUAAUA ACUUCUCGAA UCGCACGGCC UUGCGCCGGC GAUGGUUCAU UCAAAUUUC UGCCCUAUCA ACUUUCGACG GCUGGGUAUU GGCCAGCCGU GGUGACAACG GGUAACGGAG GGUUAGGGCU C GACCCCGG AGAAGGAGCC UGAGAAACGG CUACUACAUC CAAGGAAGGC AGCAGGCGCG CAAAUUACCC AAUCCCGACA CG GGGAGGU AGUGACAAUA AAUACUGAUA CAGGGCUCUU UCGGGUCUUG UAAUUGGAAU GAGUACAAUU UAAAUCCCUU AAC GAGGAA CAAUUGGAGG GCAAGUCUGG UGCCAGCAGC CGCGGUAAUU CCAGCUCCAA UAGCGUAUAU UAAAGUUGUU GAGG UUAAA AAGCUCGUAG UUGAACCUUG GGCCUAGCCC GCCGGUCCGC CUCACCGCGU GCACUGGCUG GGCUGGGCCU UUCCU UCUG GAGAACCGCA UGCCCUUCAC UGGGUGUGUC GGGGAACCAG GACUUUUACU CUGAACAAAU UAGAUCGCUU AAAGAA GGC CUAUGCUUGA AUACAUUAGC AUGGAAUAAU AGAAUAGGAC GUGUGGUUCU AUUUUGUUGG UUUCUAGGAC CGCCGUA AU GAUUAAUAGG GACAGUCGGG GGCGUCAGUA UUCAAUUGUC AGAGGUGAAA UUCUUGGAUU UAUUGAAGAC UAACAACU G CGAAAGCAUU CGCCAAGGAU GUUUUCAUUA AUCAGGAACG AAAGUUAGGG GAUCGAAGAC GAUCAGAUAC CGUCGUAGU CUUAACCAUA AACUAUGCCG AUUAGGGAUC GGACGGCGUU UAUUUUUGAC CCGUUCGGCA CCUUACGAUA AAUCAAAAUG UUUGGGCUC CUGGGGGAGU AUGGUCGCAA GGCUGAAACU UAAAGAAAUU GACGGAAGGG CACCACCAGG GGUGGAGCCU G CGGCUUAA UUUGACUCAA CACGGGGAAA CUCACCAGGU CCAGACACGA UGAGGAUUGA CAGAUUGAGA GCUCUUUCUU GA UUUCGUG GGUGGUGGUG CAUGGCCGUU CUUAGUUGGU GGAGUGAUUU GUCUGCUUAA UUGCGAUAAC GAACGAGACC UUA ACCUGC UAAAUAGCCC GUAUUGCCUU GGCAGUACGC CGGCUUCUUA GAGGGACUAU CGGCUCAAGC CGAUGGAAGU UUGA GGCAA UAACAGGUCU GUGAUGCCCU UAGAUGUUCU GGGCCGCACG CGCGCUACAC UGACAGAGCC AGCGAGUACU CCCUU GGCC GAGAGGCCCG GGUAAUCUUG UUAAACUCUG UCGUGCUGGG GAUAGAGCAU UGCAAUUAUU GCUCUUCAAC GAGGAA UCC CUAGUAAGCG CAAGUCAUCA GCUUGCGUUG AUUACGUCCC UGCCCUUUGU ACACACCGCC CGUCGCUACU ACCGAUU GA ACGGCUCAGU GAGGCUUUCG GACUGGCCCA GAGAGGUCGG CAACGACCAC UCAGGGCCGG AAAGUUAUCC AAACUCGG U CGUUUAGAGG AAGUAAAAGU CGUAACAAGG UCUCCGUAGG UGAACCUGCG GAGGGAUCAU UAAUGAGUUA CCACAACUC CCAAACCCAU UGUGAACCUA CCCUUCACGU UGCUUCGGCG GGUCGGGCGC UGUGCCGCCC CUCCGGGCGA CCCGGGGCUC CCCAAAGGC CCCGGGCCCG CCGCCCGCCG GAGGUACGCA AACUCUUGUU CAUUGUACGG CCUCUCUGAG UGAAGUACUG A AUAAGUCA |
+Macromolecule #51: U3 snoRNA
| Macromolecule | Name: U3 snoRNA / type: rna / ID: 51 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
| Molecular weight | Theoretical: 87.992648 KDa |
| Sequence | String: AGCGACAAUA CUUCAGAGAA UCAUUUCUAU AGUAGUUGUC CUCUCUUGUG UUUCCUAAAG GAGCCACAGA UCCCACCCGG GUUGAUGAA CGAGAUCCUC GGCGCCCAAC AGUGAGGUCC AUUAUUUACU CUCGCUCUAC CUGCAAAGGU GGCGGUCGCG U GCCUCGUC ...String: AGCGACAAUA CUUCAGAGAA UCAUUUCUAU AGUAGUUGUC CUCUCUUGUG UUUCCUAAAG GAGCCACAGA UCCCACCCGG GUUGAUGAA CGAGAUCCUC GGCGCCCAAC AGUGAGGUCC AUUAUUUACU CUCGCUCUAC CUGCAAAGGU GGCGGUCGCG U GCCUCGUC UCGCGGCUGU AUAGAGAGUG GCGAUGAUCU GUACCCCGGC UUUCCUGAGG UCUUGCCUUC GGGCGCUCAG GC GGGGUGG GUGUCGAUGG AAGUCUGACC GGCCAUU |
+Macromolecule #52: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 52 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 2.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-5oql: |
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller

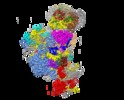





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)