[English] 日本語
 Yorodumi
Yorodumi- EMDB-11038: The mitochondrial ribosome from Tetrahymena thermophila, small su... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11038 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The mitochondrial ribosome from Tetrahymena thermophila, small subunit back protuberance mask | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Tetrahymena thermophila SB210 (eukaryote) Tetrahymena thermophila SB210 (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Tobiasson V / Amunts A | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Ciliate mitoribosome illuminates evolutionary steps of mitochondrial translation. Authors: Victor Tobiasson / Alexey Amunts /  Abstract: To understand the steps involved in the evolution of translation, we used , a ciliate with high coding capacity of the mitochondrial genome, as the model organism and characterized its mitochondrial ...To understand the steps involved in the evolution of translation, we used , a ciliate with high coding capacity of the mitochondrial genome, as the model organism and characterized its mitochondrial ribosome (mitoribosome) using cryo-EM. The structure of the mitoribosome reveals an assembly of 94-ribosomal proteins and four-rRNAs with an additional protein mass of ~700 kDa on the small subunit, while the large subunit lacks 5S rRNA. The structure also shows that the small subunit head is constrained, tRNA binding sites are formed by mitochondria-specific protein elements, conserved protein bS1 is excluded, and bacterial RNA polymerase binding site is blocked. We provide evidence for anintrinsic protein targeting system through visualization of mitochondria-specific mL105 by the exit tunnel that would facilitate the recruitment of a nascent polypeptide. Functional protein uS3m is encoded by three complementary genes from the nucleus and mitochondrion, establishing a link between genetic drift and mitochondrial translation. Finally, we reannotated nine open reading frames in the mitochondrial genome that code for mitoribosomal proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11038.map.gz emd_11038.map.gz | 580.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11038-v30.xml emd-11038-v30.xml emd-11038.xml emd-11038.xml | 38.4 KB 38.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11038.png emd_11038.png | 203.4 KB | ||
| Masks |  emd_11038_msk_1.map emd_11038_msk_1.map | 634.7 MB |  Mask map Mask map | |
| Others |  emd_11038_half_map_1.map.gz emd_11038_half_map_1.map.gz emd_11038_half_map_2.map.gz emd_11038_half_map_2.map.gz | 511.1 MB 511.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11038 http://ftp.pdbj.org/pub/emdb/structures/EMD-11038 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11038 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11038 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11038.map.gz / Format: CCP4 / Size: 634.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11038.map.gz / Format: CCP4 / Size: 634.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
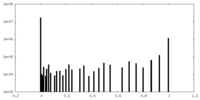
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11038_msk_1.map emd_11038_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
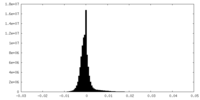
| Density Histograms |
-Half map: #2
| File | emd_11038_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
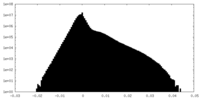
| Density Histograms |
-Half map: #1
| File | emd_11038_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
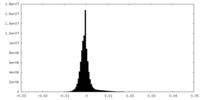
| Density Histograms |
- Sample components
Sample components
-Entire : Mitochondrial ribosome from Tetrahymena thermophila
| Entire | Name: Mitochondrial ribosome from Tetrahymena thermophila |
|---|---|
| Components |
|
-Supramolecule #1: Mitochondrial ribosome from Tetrahymena thermophila
| Supramolecule | Name: Mitochondrial ribosome from Tetrahymena thermophila / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#94 |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila SB210 (eukaryote) Tetrahymena thermophila SB210 (eukaryote) |
-Supramolecule #2: Mitochondrial ribosomal large subunit from Tetrahymena thermophila
| Supramolecule | Name: Mitochondrial ribosomal large subunit from Tetrahymena thermophila type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#48 |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila SB210 (eukaryote) Tetrahymena thermophila SB210 (eukaryote) |
-Supramolecule #3: Mitochondrial ribosomal small subunit from Tetrahymena thermophila
| Supramolecule | Name: Mitochondrial ribosomal small subunit from Tetrahymena thermophila type: complex / ID: 3 / Parent: 1 / Macromolecule list: #49-#94 |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila SB210 (eukaryote) Tetrahymena thermophila SB210 (eukaryote) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.45 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3.0 nm |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 99300 |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)