[English] 日本語
 Yorodumi
Yorodumi- EMDB-37363: CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-85... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site | |||||||||
 Map data Map data | CryoEM density map of human PI3K-alpha (P85/P110-H1047R) with Cpd2 binding at an unidentified allosteric site | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PI3K-alpha / lipid kinase / allosteric inhibition / ONCOPROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationperinuclear endoplasmic reticulum membrane / regulation of toll-like receptor 4 signaling pathway / response to muscle inactivity / phosphatidylinositol kinase activity / regulation of actin filament organization / positive regulation of focal adhesion disassembly / negative regulation of actin filament depolymerization / response to butyrate / 1-phosphatidylinositol-3-kinase regulator activity / phosphatidylinositol 3-kinase regulator activity ...perinuclear endoplasmic reticulum membrane / regulation of toll-like receptor 4 signaling pathway / response to muscle inactivity / phosphatidylinositol kinase activity / regulation of actin filament organization / positive regulation of focal adhesion disassembly / negative regulation of actin filament depolymerization / response to butyrate / 1-phosphatidylinositol-3-kinase regulator activity / phosphatidylinositol 3-kinase regulator activity / response to L-leucine / positive regulation of endoplasmic reticulum unfolded protein response / IRS-mediated signalling / phosphatidylinositol 3-kinase activator activity / T follicular helper cell differentiation / interleukin-18-mediated signaling pathway / phosphatidylinositol 3-kinase complex / PI3K events in ERBB4 signaling / phosphatidylinositol 3-kinase regulatory subunit binding / myeloid leukocyte migration / autosome genomic imprinting / neurotrophin TRKA receptor binding / cellular response to hydrostatic pressure / Activated NTRK2 signals through PI3K / regulation of cellular respiration / cis-Golgi network / negative regulation of fibroblast apoptotic process / transmembrane receptor protein tyrosine kinase adaptor activity / Activated NTRK3 signals through PI3K / ErbB-3 class receptor binding / phosphatidylinositol 3-kinase complex, class IB / negative regulation of stress fiber assembly / positive regulation of protein localization to membrane / 1-phosphatidylinositol-4-phosphate 3-kinase activity / Signaling by cytosolic FGFR1 fusion mutants / Co-stimulation by ICOS / RHOD GTPase cycle / vasculature development / cardiac muscle cell contraction / phosphatidylinositol 3-kinase complex, class IA / Nephrin family interactions / RHOF GTPase cycle / kinase activator activity / anoikis / Signaling by LTK in cancer / phosphatidylinositol-3-phosphate biosynthetic process / Signaling by LTK / positive regulation of leukocyte migration / MET activates PI3K/AKT signaling / relaxation of cardiac muscle / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / RND1 GTPase cycle / PI3K/AKT activation / RND2 GTPase cycle / phosphatidylinositol-4,5-bisphosphate 3-kinase / RND3 GTPase cycle / positive regulation of filopodium assembly / vascular endothelial growth factor signaling pathway / phosphatidylinositol 3-kinase / growth hormone receptor signaling pathway / insulin binding / Signaling by ALK / 1-phosphatidylinositol-3-kinase activity / RHOV GTPase cycle / RHOB GTPase cycle / natural killer cell mediated cytotoxicity / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / PI-3K cascade:FGFR3 / GP1b-IX-V activation signalling / negative regulation of macroautophagy / response to dexamethasone / PI-3K cascade:FGFR2 / phosphatidylinositol-mediated signaling / PI-3K cascade:FGFR4 / PI-3K cascade:FGFR1 / RHOC GTPase cycle / RHOJ GTPase cycle / negative regulation of osteoclast differentiation / phosphatidylinositol phosphate biosynthetic process / Synthesis of PIPs at the plasma membrane / intracellular glucose homeostasis / RHOU GTPase cycle / CDC42 GTPase cycle / RET signaling / negative regulation of anoikis / insulin receptor substrate binding / Interleukin-3, Interleukin-5 and GM-CSF signaling / PI3K events in ERBB2 signaling / intercalated disc / PI3K Cascade / T cell differentiation / negative regulation of cell-matrix adhesion / RHOG GTPase cycle / extrinsic apoptotic signaling pathway via death domain receptors / CD28 dependent PI3K/Akt signaling / regulation of multicellular organism growth / Role of LAT2/NTAL/LAB on calcium mobilization / RHOA GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
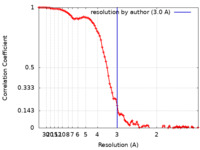
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Huang X / Ren X / Zhong W | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Cryo-EM structures reveal two allosteric inhibition modes of PI3Kα involving a re-shaping of the activation loop. Authors: Xiuliang Huang / Kailiang Wang / Jing Han / Xiumei Chen / Zhenglin Wang / Tianlun Wu / Bo Yu / Feng Zhao / Xinjuan Wang / Huijuan Li / Zhi Xie / Xiaotian Zhu / Wenge Zhong / Xiaoming Ren /   Abstract: PI3Kα is a lipid kinase that phosphorylates PIP2 and generates PIP3. The hyperactive PI3Kα mutation, H1047R, accounts for about 14% of breast cancer, making it a highly attractive target for drug ...PI3Kα is a lipid kinase that phosphorylates PIP2 and generates PIP3. The hyperactive PI3Kα mutation, H1047R, accounts for about 14% of breast cancer, making it a highly attractive target for drug discovery. Here, we report the cryo-EM structures of PI3Kα bound to two different allosteric inhibitors QR-7909 and QR-8557 at a global resolution of 2.7 Å and 3.0 Å, respectively. The structures reveal two distinct binding pockets on the opposite sides of the activation loop. Structural and MD simulation analyses show that the allosteric binding of QR-7909 and QR-8557 inhibit PI3Kα hyper-activity by reducing the fluctuation and mobility of the activation loop. Our work provides a strong rational basis for a further optimization and development of highly selective drug candidates to treat PI3Kα-driven cancers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37363.map.gz emd_37363.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37363-v30.xml emd-37363-v30.xml emd-37363.xml emd-37363.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37363_fsc.xml emd_37363_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_37363.png emd_37363.png | 45.4 KB | ||
| Filedesc metadata |  emd-37363.cif.gz emd-37363.cif.gz | 6.3 KB | ||
| Others |  emd_37363_half_map_1.map.gz emd_37363_half_map_1.map.gz emd_37363_half_map_2.map.gz emd_37363_half_map_2.map.gz | 49.7 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37363 http://ftp.pdbj.org/pub/emdb/structures/EMD-37363 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37363 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37363 | HTTPS FTP |
-Related structure data
| Related structure data |  8w9bMC  8w9aC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37363.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37363.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM density map of human PI3K-alpha (P85/P110-H1047R) with Cpd2 binding at an unidentified allosteric site | ||||||||||||||||||||||||||||||||||||
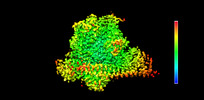
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.74 Å | ||||||||||||||||||||||||||||||||||||
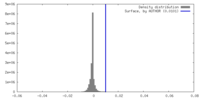
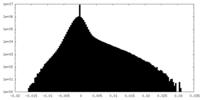
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1 of Cpd2-bound human PI3K-alpha (P85/P110-H1047R)
| File | emd_37363_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of Cpd2-bound human PI3K-alpha (P85/P110-H1047R) | ||||||||||||
| Projections & Slices |
| ||||||||||||
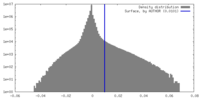
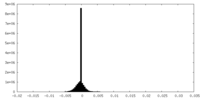
| Density Histograms |
-Half map: Half map 2 of Cpd2-bound human PI3K-alpha (P85/P110-H1047R)
| File | emd_37363_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of Cpd2-bound human PI3K-alpha (P85/P110-H1047R) | ||||||||||||
| Projections & Slices |
| ||||||||||||
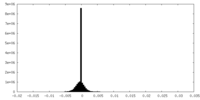
| Density Histograms |
- Sample components
Sample components
-Entire : human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an all...
| Entire | Name: human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site |
|---|---|
| Components |
|
-Supramolecule #1: human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an all...
| Supramolecule | Name: human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit ...
| Macromolecule | Name: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: phosphatidylinositol 3-kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124.23075 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MPPRPSSGEL WGIHLMPPRI LVECLLPNGM IVTLECLREA TLITIKHELF KEARKYPLHQ LLQDESSYIF VSVTQEAERE EFFDETRRL CDLRLFQPFL KVIEPVGNRE EKILNREIGF AIGMPVCEFD MVKDPEVQDF RRNILNVCKE AVDLRDLNSP H SRAMYVYP ...String: MPPRPSSGEL WGIHLMPPRI LVECLLPNGM IVTLECLREA TLITIKHELF KEARKYPLHQ LLQDESSYIF VSVTQEAERE EFFDETRRL CDLRLFQPFL KVIEPVGNRE EKILNREIGF AIGMPVCEFD MVKDPEVQDF RRNILNVCKE AVDLRDLNSP H SRAMYVYP PNVESSPELP KHIYNKLDKG QIIVVIWVIV SPNNDKQKYT LKINHDCVPE QVIAEAIRKK TRSMLLSSEQ LK LCVLEYQ GKYILKVCGC DEYFLEKYPL SQYKYIRSCI MLGRMPNLML MAKESLYSQL PMDCFTMPSY SRRISTATPY MNG ETSTKS LWVINSALRI KILCATYVNV NIRDIDKIYV RTGIYHGGEP LCDNVNTQRV PCSNPRWNEW LNYDIYIPDL PRAA RLCLS ICSVKGRKGA KEEHCPLAWG NINLFDYTDT LVSGKMALNL WPVPHGLEDL LNPIGVTGSN PNKETPCLEL EFDWF SSVV KFPDMSVIEE HANWSVSREA GFSYSHAGLS NRLARDNELR ENDKEQLKAI STRDPLSEIT EQEKDFLWSH RHYCVT IPE ILPKLLLSVK WNSRDEVAQM YCLVKDWPPI KPEQAMELLD CNYPDPMVRG FAVRCLEKYL TDDKLSQYLI QLVQVLK YE QYLDNLLVRF LLKKALTNQR IGHFFFWHLK SEMHNKTVSQ RFGLLLESYC RACGMYLKHL NRQVEAMEKL INLTDILK Q EKKDETQKVQ MKFLVEQMRR PDFMDALQGF LSPLNPAHQL GNLRLEECRI MSSAKRPLWL NWENPDIMSE LLFQNNEII FKNGDDLRQD MLTLQIIRIM ENIWQNQGLD LRMLPYGCLS IGDCVGLIEV VRNSHTIMQI QCKGGLKGAL QFNSHTLHQW LKDKNKGEI YDAAIDLFTR SCAGYCVATF ILGIGDRHNS NIMVKDDGQL FHIDFGHFLD HKKKKFGYKR ERVPFVLTQD F LIVISKGA QECTKTREFE RFQEMCYKAY LAIRQHANLF INLFSMMLGS GMPELQSFDD IAYIRKTLAL DKTEQEALEY FM KQMNDAR HGGWTTKMDA AAHTIKQHAL N UniProtKB: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform |
-Macromolecule #2: Phosphatidylinositol 3-kinase regulatory subunit alpha
| Macromolecule | Name: Phosphatidylinositol 3-kinase regulatory subunit alpha type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.666961 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MNNNMSLQDA EWYWGDISRE EVNEKLRDTA DGTFLVRDAS TKMHGDYTLT LRKGGNNKLI KIFHRDGKYG FSDPLTFSSV VELINHYRN ESLAQYNPKL DVKLLYPVSK YQQDQVVKED NIEAVGKKLH EYNTQFQEKS REYDRLYEEY TRTSQEIQMK R TAIEAFNE ...String: MNNNMSLQDA EWYWGDISRE EVNEKLRDTA DGTFLVRDAS TKMHGDYTLT LRKGGNNKLI KIFHRDGKYG FSDPLTFSSV VELINHYRN ESLAQYNPKL DVKLLYPVSK YQQDQVVKED NIEAVGKKLH EYNTQFQEKS REYDRLYEEY TRTSQEIQMK R TAIEAFNE TIKIFEEQCQ TQERYSKEYI EKFKREGNEK EIQRIMHNYD KLKSRISEII DSRRRLEEDL KKQAAEYREI DK RMNSIKP DLIQLRKTRD QYLMWLTQKG VRQKKLNEWL GN UniProtKB: Phosphatidylinositol 3-kinase regulatory subunit alpha |
-Macromolecule #3: 1-[(1S)-1-(5-fluoranyl-3-methyl-1-benzofuran-2-yl)-2-methyl-propy...
| Macromolecule | Name: 1-[(1S)-1-(5-fluoranyl-3-methyl-1-benzofuran-2-yl)-2-methyl-propyl]-3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)urea type: ligand / ID: 3 / Number of copies: 1 / Formula: UJ3 |
|---|---|
| Molecular weight | Theoretical: 395.427 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 48.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)