+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3690 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
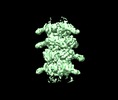
| Title | Empty T5 tail | |||||||||
 Map data Map data | Empty T5 tail | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | Arnaud C / Effantin G / Vives C / Engilberge S / Bacia M / Boulanger P / Girard E / Schoehn G / Breyton C | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection. Authors: Charles-Adrien Arnaud / Grégory Effantin / Corinne Vivès / Sylvain Engilberge / Maria Bacia / Pascale Boulanger / Eric Girard / Guy Schoehn / Cécile Breyton /  Abstract: The vast majority of phages, bacterial viruses, possess a tail ensuring host recognition, cell wall perforation and safe viral DNA transfer from the capsid to the host cytoplasm. Long flexible tails ...The vast majority of phages, bacterial viruses, possess a tail ensuring host recognition, cell wall perforation and safe viral DNA transfer from the capsid to the host cytoplasm. Long flexible tails are formed from the tail tube protein (TTP) polymerised as hexameric rings around and stacked along the tape measure protein (TMP). Here, we report the crystal structure of T5 TTP pb6 at 2.2 Å resolution. Pb6 is unusual in forming a trimeric ring, although structure analysis reveals homology with all classical TTPs and related tube proteins of bacterial puncturing devices (type VI secretion system and R-pyocin). Structures of T5 tail tubes before and after interaction with the host receptor were determined by cryo-electron microscopy at 6 Å resolution. Comparison of these two structures reveals that host-binding information is not propagated to the capsid through conformational changes in the tail tube, suggesting a role of the TMP in this information transduction process. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3690.map.gz emd_3690.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3690-v30.xml emd-3690-v30.xml emd-3690.xml emd-3690.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3690.png emd_3690.png | 76.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3690 http://ftp.pdbj.org/pub/emdb/structures/EMD-3690 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3690 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3690 | HTTPS FTP |
-Validation report
| Summary document |  emd_3690_validation.pdf.gz emd_3690_validation.pdf.gz | 244 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3690_full_validation.pdf.gz emd_3690_full_validation.pdf.gz | 243.1 KB | Display | |
| Data in XML |  emd_3690_validation.xml.gz emd_3690_validation.xml.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3690 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3690 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3690 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3690 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3690.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3690.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Empty T5 tail | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.64 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Empty T5 tail
| Entire | Name: Empty T5 tail |
|---|---|
| Components |
|
-Supramolecule #1: Empty T5 tail
| Supramolecule | Name: Empty T5 tail / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 40.6 Å Applied symmetry - Helical parameters - Δ&Phi: 39.1 ° Applied symmetry - Helical parameters - Axial symmetry: C3 (3 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 5.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 22704 |
|---|---|
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















