+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4788 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | D. melanogaster CMG-DNA with ATP, State 2B | |||||||||
 Map data Map data | State 2B, sharpened map (RELION PostProcess) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helicase / ATPase / AAA+ / DNA unwinding / hydrolase | |||||||||
| Function / homology |  Function and homology information Function and homology informationUnwinding of DNA / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / DNA endoreduplication / Activation of ATR in response to replication stress / Activation of the pre-replicative complex / eggshell chorion gene amplification / Orc1 removal from chromatin / DNA amplification / DNA strand elongation involved in mitotic DNA replication ...Unwinding of DNA / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / DNA endoreduplication / Activation of ATR in response to replication stress / Activation of the pre-replicative complex / eggshell chorion gene amplification / Orc1 removal from chromatin / DNA amplification / DNA strand elongation involved in mitotic DNA replication / GINS complex / mitotic DNA replication preinitiation complex assembly / resolution of meiotic recombination intermediates / premeiotic DNA replication / mitotic DNA replication / CMG complex / DNA replication preinitiation complex / MCM complex / double-strand break repair via break-induced replication / mitotic DNA replication initiation / chromosome condensation / DNA strand elongation involved in DNA replication / DNA replication origin binding / DNA replication initiation / DNA helicase activity / regulation of DNA-templated transcription elongation / mitotic spindle organization / meiotic cell cycle / helicase activity / mitotic cell cycle / single-stranded DNA binding / DNA helicase / DNA replication / cell division / chromatin binding / ATP hydrolysis activity / zinc ion binding / ATP binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
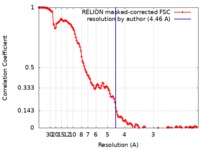
| Method | single particle reconstruction / cryo EM / Resolution: 4.46 Å | |||||||||
 Authors Authors | Eickhoff P / Martino F / Locke J / Nans A / Costa A | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2019 Journal: Cell Rep / Year: 2019Title: Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome. Authors: Patrik Eickhoff / Hazal B Kose / Fabrizio Martino / Tatjana Petojevic / Ferdos Abid Ali / Julia Locke / Nele Tamberg / Andrea Nans / James M Berger / Michael R Botchan / Hasan Yardimci / Alessandro Costa /    Abstract: In the eukaryotic replisome, DNA unwinding by the Cdc45-MCM-Go-Ichi-Ni-San (GINS) (CMG) helicase requires a hexameric ring-shaped ATPase named minichromosome maintenance (MCM), which spools single- ...In the eukaryotic replisome, DNA unwinding by the Cdc45-MCM-Go-Ichi-Ni-San (GINS) (CMG) helicase requires a hexameric ring-shaped ATPase named minichromosome maintenance (MCM), which spools single-stranded DNA through its central channel. Not all six ATPase sites are required for unwinding; however, the helicase mechanism is unknown. We imaged ATP-hydrolysis-driven translocation of the CMG using cryo-electron microscopy (cryo-EM) and found that the six MCM subunits engage DNA using four neighboring protomers at a time, with ATP binding promoting DNA engagement. Morphing between different helicase states leads us to suggest a non-symmetric hand-over-hand rotary mechanism, explaining the asymmetric requirements of ATPase function around the MCM ring of the CMG. By imaging of a higher-order replisome assembly, we find that the Mrc1-Csm3-Tof1 fork-stabilization complex strengthens the interaction between parental duplex DNA and the CMG at the fork, which might support the coupling between DNA translocation and fork unwinding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4788.map.gz emd_4788.map.gz | 14.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4788-v30.xml emd-4788-v30.xml emd-4788.xml emd-4788.xml | 43.4 KB 43.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4788_fsc.xml emd_4788_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_4788.png emd_4788.png | 133.9 KB | ||
| Filedesc metadata |  emd-4788.cif.gz emd-4788.cif.gz | 11.3 KB | ||
| Others |  emd_4788_additional.map.gz emd_4788_additional.map.gz emd_4788_half_map_1.map.gz emd_4788_half_map_1.map.gz emd_4788_half_map_2.map.gz emd_4788_half_map_2.map.gz | 171.7 MB 171.2 MB 171.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4788 http://ftp.pdbj.org/pub/emdb/structures/EMD-4788 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4788 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4788 | HTTPS FTP |
-Related structure data
| Related structure data |  6razMC  4785C  4786C  4787C  6rawC  6raxC  6rayC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4788.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4788.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | State 2B, sharpened map (RELION PostProcess) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
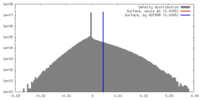
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: State 2B, non-sharpened map (RELION Refine3D)
| File | emd_4788_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | State 2B, non-sharpened map (RELION Refine3D) | ||||||||||||
| Projections & Slices |
| ||||||||||||
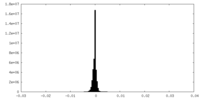
| Density Histograms |
-Half map: State 2B, half-map 2 (RELION Refine3D)
| File | emd_4788_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | State 2B, half-map 2 (RELION Refine3D) | ||||||||||||
| Projections & Slices |
| ||||||||||||
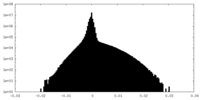
| Density Histograms |
-Half map: State 2B, half-map 1 (RELION Refine3D)
| File | emd_4788_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | State 2B, half-map 1 (RELION Refine3D) | ||||||||||||
| Projections & Slices |
| ||||||||||||
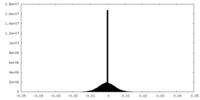
| Density Histograms |
- Sample components
Sample components
+Entire : CMG helicase bound to forked DNA in the presence of ATP, State 2B
+Supramolecule #1: CMG helicase bound to forked DNA in the presence of ATP, State 2B
+Supramolecule #2: CMG helicase
+Supramolecule #3: DNA
+Macromolecule #1: DNA
+Macromolecule #2: DNA
+Macromolecule #3: CDC45L
+Macromolecule #4: IP07275p
+Macromolecule #5: Probable DNA replication complex GINS protein PSF2
+Macromolecule #6: AT18545p
+Macromolecule #7: DNA replication complex GINS protein SLD5
+Macromolecule #8: DNA replication licensing factor Mcm2
+Macromolecule #9: DNA replication licensing factor Mcm5
+Macromolecule #10: DNA replication licensing factor Mcm6
+Macromolecule #11: DNA replication licensing factor Mcm3
+Macromolecule #12: DNA replication licensing factor MCM4
+Macromolecule #13: DNA replication licensing factor Mcm7
+Macromolecule #14: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #15: ADENOSINE-5'-TRIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)