+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vgamma5 Vdelta1 TCR complex (MPDI/TMDI) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Receptor / IMMUNE SYSTEM | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / Resolution: 3.9 Å | |||||||||
 Authors Authors | Xin W / Huang B / Chi X / Liu Y / Xu M / Zhang Y / Li X / Su Q / Zhou Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structures of human γδ T cell receptor-CD3 complex. Authors: Weizhi Xin / Bangdong Huang / Ximin Chi / Yuehua Liu / Mengjiao Xu / Yuanyuan Zhang / Xu Li / Qiang Su / Qiang Zhou /  Abstract: Gamma delta (γδ) T cells, a unique T cell subgroup, are crucial in various immune responses and immunopathology. The γδ T cell receptor (TCR), which is generated by γδ T cells, recognizes a ...Gamma delta (γδ) T cells, a unique T cell subgroup, are crucial in various immune responses and immunopathology. The γδ T cell receptor (TCR), which is generated by γδ T cells, recognizes a diverse range of antigens independently of the major histocompatibility complex. The γδ TCR associates with CD3 subunits, initiating T cell activation and holding great potential in immunotherapy. Here we report the structures of two prototypical human Vγ9Vδ2 and Vγ5Vδ1 TCR-CD3 complexes, revealing two distinct assembly mechanisms that depend on Vγ usage. The Vγ9Vδ2 TCR-CD3 complex is monomeric, with considerable conformational flexibility in the TCRγ-TCRδ extracellular domain and connecting peptides. The length of the connecting peptides regulates the ligand association and T cell activation. A cholesterol-like molecule wedges into the transmembrane region, exerting an inhibitory role in TCR signalling. The Vγ5Vδ1 TCR-CD3 complex displays a dimeric architecture, whereby two protomers nestle back to back through the Vγ5 domains of the TCR extracellular domains. Our biochemical and biophysical assays further corroborate the dimeric structure. Importantly, the dimeric form of the Vγ5Vδ1 TCR is essential for T cell activation. These findings reveal organizing principles of the γδ TCR-CD3 complex, providing insights into the unique properties of γδ TCR and facilitating immunotherapeutic interventions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36152.map.gz emd_36152.map.gz | 31.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36152-v30.xml emd-36152-v30.xml emd-36152.xml emd-36152.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36152.png emd_36152.png | 37.5 KB | ||
| Filedesc metadata |  emd-36152.cif.gz emd-36152.cif.gz | 3.8 KB | ||
| Others |  emd_36152_half_map_1.map.gz emd_36152_half_map_1.map.gz emd_36152_half_map_2.map.gz emd_36152_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36152 http://ftp.pdbj.org/pub/emdb/structures/EMD-36152 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36152 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36152 | HTTPS FTP |
-Validation report
| Summary document |  emd_36152_validation.pdf.gz emd_36152_validation.pdf.gz | 822.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36152_full_validation.pdf.gz emd_36152_full_validation.pdf.gz | 821.8 KB | Display | |
| Data in XML |  emd_36152_validation.xml.gz emd_36152_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_36152_validation.cif.gz emd_36152_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36152 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36152 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36152 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36152 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36152.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36152.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36152_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
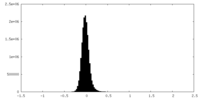
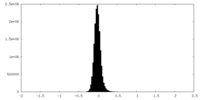
| Density Histograms |
-Half map: #2
| File | emd_36152_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
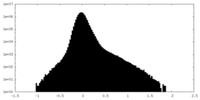
| Density Histograms |
- Sample components
Sample components
-Entire : Vgamma5 Vdelta1 TCR complex (MPDI/TMDI)
| Entire | Name: Vgamma5 Vdelta1 TCR complex (MPDI/TMDI) |
|---|---|
| Components |
|
-Supramolecule #1: Vgamma5 Vdelta1 TCR complex (MPDI/TMDI)
| Supramolecule | Name: Vgamma5 Vdelta1 TCR complex (MPDI/TMDI) / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
 Processing Processing | single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 100364 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)