[English] 日本語
 Yorodumi
Yorodumi- EMDB-3572: Localized reconstruction of bacteriophage phi6 packaging hexamer P4 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3572 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Localized reconstruction of bacteriophage phi6 packaging hexamer P4 | |||||||||
 Map data Map data | Localized reconstruction of bacteriophage phi6 packaging hexamer P4 in situ | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | packaging / ATPase / vertex / hyrdolase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral procapsid / viral genome packaging / ribonucleoside triphosphate phosphatase activity / viral capsid / nucleoside-triphosphate phosphatase / ATP binding Similarity search - Function | |||||||||
| Biological species |  Pseudomonas phage phi6 (virus) Pseudomonas phage phi6 (virus) | |||||||||
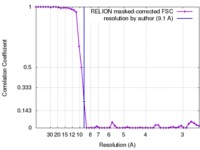
| Method | single particle reconstruction / cryo EM / Resolution: 9.1 Å | |||||||||
 Authors Authors | Sun Z / El Omari K | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2013 Journal: Nucleic Acids Res / Year: 2013Title: Tracking in atomic detail the functional specializations in viral RecA helicases that occur during evolution. Authors: Kamel El Omari / Christoph Meier / Denis Kainov / Geoff Sutton / Jonathan M Grimes / Minna M Poranen / Dennis H Bamford / Roman Tuma / David I Stuart / Erika J Mancini /  Abstract: Many complex viruses package their genomes into empty protein shells and bacteriophages of the Cystoviridae family provide some of the simplest models for this. The cystoviral hexameric NTPase, P4, ...Many complex viruses package their genomes into empty protein shells and bacteriophages of the Cystoviridae family provide some of the simplest models for this. The cystoviral hexameric NTPase, P4, uses chemical energy to translocate single-stranded RNA genomic precursors into the procapsid. We previously dissected the mechanism of RNA translocation for one such phage, 12, and have now investigated three further highly divergent, cystoviral P4 NTPases (from 6, 8 and 13). High-resolution crystal structures of the set of P4s allow a structure-based phylogenetic analysis, which reveals that these proteins form a distinct subfamily of the RecA-type ATPases. Although the proteins share a common catalytic core, they have different specificities and control mechanisms, which we map onto divergent N- and C-terminal domains. Thus, the RNA loading and tight coupling of NTPase activity with RNA translocation in 8 P4 is due to a remarkable C-terminal structure, which wraps right around the outside of the molecule to insert into the central hole where RNA binds to coupled L1 and L2 loops, whereas in 12 P4, a C-terminal residue, serine 282, forms a specific hydrogen bond to the N7 of purines ring to confer purine specificity for the 12 enzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3572.map.gz emd_3572.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3572-v30.xml emd-3572-v30.xml emd-3572.xml emd-3572.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3572_fsc.xml emd_3572_fsc.xml | 4.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3572.png emd_3572.png | 172.4 KB | ||
| Filedesc metadata |  emd-3572.cif.gz emd-3572.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3572 http://ftp.pdbj.org/pub/emdb/structures/EMD-3572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3572 | HTTPS FTP |
-Related structure data
| Related structure data |  5muvMC  3571C  3573C  5muuC  5muwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3572.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3572.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Localized reconstruction of bacteriophage phi6 packaging hexamer P4 in situ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pseudomonas phage phi6
| Entire | Name:  Pseudomonas phage phi6 (virus) Pseudomonas phage phi6 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas phage phi6
| Supramolecule | Name: Pseudomonas phage phi6 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 10879 / Sci species name: Pseudomonas phage phi6 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Pseudomonas syringae (bacteria) Pseudomonas syringae (bacteria) |
-Macromolecule #1: Packaging enzyme P4
| Macromolecule | Name: Packaging enzyme P4 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: nucleoside-triphosphate phosphatase |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phi6 (virus) Pseudomonas phage phi6 (virus) |
| Molecular weight | Theoretical: 32.67874 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPIVVTQAHI DRVGIAADLL DASPVSLQVL GRPTAINTVV IKTYIAAVME LASKQGGSLA GVDIRPSVLL KDTAIFTKPK AKSADVESD VDVLDTGIYS VPGLARKPVT HRWPSEGIYS GVTALMGATG SGKSITLNEK LRPDVLIRWG EVAEAYDELD T AVHISTLD ...String: MPIVVTQAHI DRVGIAADLL DASPVSLQVL GRPTAINTVV IKTYIAAVME LASKQGGSLA GVDIRPSVLL KDTAIFTKPK AKSADVESD VDVLDTGIYS VPGLARKPVT HRWPSEGIYS GVTALMGATG SGKSITLNEK LRPDVLIRWG EVAEAYDELD T AVHISTLD EMLIVCIGLG ALGFNVAVDS VRPLLFRLKG AASAGGIVAV FYSLLTDISN LFTQYDCSVV MVVNPMVDAE KI EYVFGQV MASTVGAILC ADGNVSRTMF RTNKGRIFNG AAPLAADTHM PSMDRPTSMK ALDHTSIASV AP UniProtKB: Packaging enzyme P4 |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Grid | Model: C-flat / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80.0 K / Max: 120.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-22 / Number real images: 900 / Average exposure time: 0.2 sec. / Average electron dose: 0.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.3 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-5muv: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)