+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ligase / complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of mitotic cell cycle phase transition / trophectodermal cellular morphogenesis / liver morphogenesis / POZ domain binding / perinuclear endoplasmic reticulum membrane / regulation of toll-like receptor 4 signaling pathway / nuclear protein quality control by the ubiquitin-proteasome system / polar microtubule / COPII vesicle coating / phosphatidylinositol kinase activity ...positive regulation of mitotic cell cycle phase transition / trophectodermal cellular morphogenesis / liver morphogenesis / POZ domain binding / perinuclear endoplasmic reticulum membrane / regulation of toll-like receptor 4 signaling pathway / nuclear protein quality control by the ubiquitin-proteasome system / polar microtubule / COPII vesicle coating / phosphatidylinositol kinase activity / anaphase-promoting complex-dependent catabolic process / phosphatidylinositol 3-kinase regulator activity / positive regulation of focal adhesion disassembly / IRS-mediated signalling / phosphatidylinositol 3-kinase activator activity / 1-phosphatidylinositol-3-kinase regulator activity / positive regulation of endoplasmic reticulum unfolded protein response / interleukin-18-mediated signaling pathway / myeloid leukocyte migration / phosphatidylinositol 3-kinase complex / T follicular helper cell differentiation / PI3K events in ERBB4 signaling / regulation protein catabolic process at postsynapse / cullin-RING-type E3 NEDD8 transferase / phosphatidylinositol 3-kinase regulatory subunit binding / RHOBTB3 ATPase cycle / NEDD8 transferase activity / cell projection organization / neurotrophin TRKA receptor binding / cullin-RING ubiquitin ligase complex / positive regulation of mitotic metaphase/anaphase transition / Activated NTRK2 signals through PI3K / embryonic cleavage / cellular response to chemical stress / Cul7-RING ubiquitin ligase complex / cis-Golgi network / Activated NTRK3 signals through PI3K / ErbB-3 class receptor binding / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / transmembrane receptor protein tyrosine kinase adaptor activity / stem cell division / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / Signaling by cytosolic FGFR1 fusion mutants / Co-stimulation by ICOS / RHOD GTPase cycle / positive regulation of protein autoubiquitination / RHOF GTPase cycle / protein neddylation / phosphatidylinositol 3-kinase complex, class IA / kinase activator activity / Nephrin family interactions / Signaling by LTK in cancer / Notch binding / positive regulation of leukocyte migration / Signaling by LTK / NEDD8 ligase activity / fibroblast apoptotic process / MET activates PI3K/AKT signaling / PI3K/AKT activation / negative regulation of response to oxidative stress / RHOBTB1 GTPase cycle / negative regulation of stress fiber assembly / RND1 GTPase cycle / Cul5-RING ubiquitin ligase complex / positive regulation of filopodium assembly / RND2 GTPase cycle / RND3 GTPase cycle / insulin binding / SCF ubiquitin ligase complex / growth hormone receptor signaling pathway / Cul2-RING ubiquitin ligase complex / Cul4A-RING E3 ubiquitin ligase complex / ubiquitin-ubiquitin ligase activity / Signaling by ALK / negative regulation of type I interferon production / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / Cul3-RING ubiquitin ligase complex / RHOV GTPase cycle / PI-3K cascade:FGFR3 / RHOB GTPase cycle / mitotic metaphase chromosome alignment / natural killer cell mediated cytotoxicity / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / GP1b-IX-V activation signalling / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / PI-3K cascade:FGFR2 / PI-3K cascade:FGFR4 / negative regulation of mitophagy / negative regulation of Rho protein signal transduction / PI-3K cascade:FGFR1 / stress fiber assembly / Prolactin receptor signaling / RHOC GTPase cycle / RHOJ GTPase cycle / positive regulation of cytokinesis / intracellular glucose homeostasis / phosphatidylinositol phosphate biosynthetic process / negative regulation of osteoclast differentiation / Synthesis of PIPs at the plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Hu Y / Mao Q / Chen Z / Sun L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3. Authors: Yuxia Hu / Zhao Zhang / Qiyu Mao / Xiang Zhang / Aihua Hao / Yu Xun / Yeda Wang / Lin Han / Wuqiang Zhan / Qianying Liu / Yue Yin / Chao Peng / Eva Marie Y Moresco / Zhenguo Chen / Bruce Beutler / Lei Sun /   Abstract: Phosphatidylinositol 3-kinase α, a heterodimer of catalytic p110α and one of five regulatory subunits, mediates insulin- and insulin like growth factor-signaling and, frequently, oncogenesis. ...Phosphatidylinositol 3-kinase α, a heterodimer of catalytic p110α and one of five regulatory subunits, mediates insulin- and insulin like growth factor-signaling and, frequently, oncogenesis. Cellular levels of the regulatory p85α subunit are tightly controlled by regulated proteasomal degradation. In adipose tissue and growth plates, failure of K48-linked p85α ubiquitination causes diabetes, lipodystrophy and dwarfism in mice, as in humans with SHORT syndrome. Here we elucidated the structures of the key ubiquitin ligase complexes regulating p85α availability. Specificity is provided by the substrate receptor KBTBD2, which recruits p85α to the cullin3-RING E3 ubiquitin ligase (CRL3). CRL3 forms multimers, which disassemble into dimers upon substrate binding (CRL3-p85α) and/or neddylation by the activator NEDD8 (CRL3~N8), leading to p85α ubiquitination and degradation. Deactivation involves dissociation of NEDD8 mediated by the COP9 signalosome and displacement of KBTBD2 by the inhibitor CAND1. The hereby identified structural basis of p85α regulation opens the way to better understanding disturbances of glucose regulation, growth and cancer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34452.map.gz emd_34452.map.gz | 193.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34452-v30.xml emd-34452-v30.xml emd-34452.xml emd-34452.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34452.png emd_34452.png | 61.3 KB | ||
| Filedesc metadata |  emd-34452.cif.gz emd-34452.cif.gz | 7.3 KB | ||
| Others |  emd_34452_additional_1.map.gz emd_34452_additional_1.map.gz emd_34452_additional_2.map.gz emd_34452_additional_2.map.gz emd_34452_additional_3.map.gz emd_34452_additional_3.map.gz emd_34452_half_map_1.map.gz emd_34452_half_map_1.map.gz emd_34452_half_map_2.map.gz emd_34452_half_map_2.map.gz | 192.4 MB 200.2 MB 200.2 MB 170.9 MB 171.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34452 http://ftp.pdbj.org/pub/emdb/structures/EMD-34452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34452 | HTTPS FTP |
-Related structure data
| Related structure data |  8h36MC  8gq6C  8h33C  8h34C  8h35C  8h37C  8h38C  8h3aC  8h3fC  8h3qC  8h3rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34452.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34452.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.332 Å | ||||||||||||||||||||||||||||||||||||
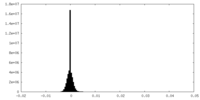
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #3
| File | emd_34452_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
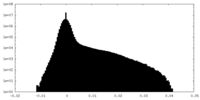
| Density Histograms |
-Additional map: #2
| File | emd_34452_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_34452_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34452_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34452_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : KBTBD2-CUL3-Rbx1-p85a dimeric complex
| Entire | Name: KBTBD2-CUL3-Rbx1-p85a dimeric complex |
|---|---|
| Components |
|
-Supramolecule #1: KBTBD2-CUL3-Rbx1-p85a dimeric complex
| Supramolecule | Name: KBTBD2-CUL3-Rbx1-p85a dimeric complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: E3 ubiquitin-protein ligase RBX1
| Macromolecule | Name: E3 ubiquitin-protein ligase RBX1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.289977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAAMDVDTP SGTNSGAGKK RFEVKKWNAV ALWAWDIVVD NCAICRNHIM DLCIECQANQ ASATSEECTV AWGVCNHAFH FHCISRWLK TRQVCPLDNR EWEFQKYGH UniProtKB: E3 ubiquitin-protein ligase RBX1 |
-Macromolecule #2: Kelch repeat and BTB domain-containing protein 2
| Macromolecule | Name: Kelch repeat and BTB domain-containing protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.431375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTQDERQIN TEYAVSLLEQ LKLFYEQQLF TDIVLIVEGT EFPCHKMVLA TCSSYFRAMF MSGLSESKQT HVHLRNVDAA TLQIIITYA YTGNLAMNDS TVEQLYETAC FLQVEDVLQR CREYLIKKIN AENCVRLLSF ADLFSCEELK QSAKRMVEHK F TAVYHQDA ...String: MSTQDERQIN TEYAVSLLEQ LKLFYEQQLF TDIVLIVEGT EFPCHKMVLA TCSSYFRAMF MSGLSESKQT HVHLRNVDAA TLQIIITYA YTGNLAMNDS TVEQLYETAC FLQVEDVLQR CREYLIKKIN AENCVRLLSF ADLFSCEELK QSAKRMVEHK F TAVYHQDA FMQLSHDLLI DILSSDNLNV EKEETVREAA MLWLEYNTES RSQYLSSVLS QIRIDALSEV TQRAWFQGLP PN DKSVVVQ GLYKDMPKFF KPRLGMTKEE MMIFIEASSE NPCSLYSSVC YSPQAEKVYK LCSPPADLHK VGTVVTPDND IYI AGGQVP LKNTKTNHSK TSKLQTAFRT VNCFYWFDAQ QNTWFPKTPM LFVRIKPSLV CCEGYIYAIG GDSVGGELNR RTVE RYDTE KDEWTMVSPL PCAWQWSAAV VVHDCIYVMT LNLMYCYFPR SDSWVEMAMR QTSRSFASAA AFGDKIFYIG GLHIA TNSG IRLPSGTVDG SSVTVEIYDV NKNEWKMAAN IPAKRYSDPC VRAVVISNSL CVFMRETHLN ERAKYVTYQY DLELDR WSL RQHISERVLW DLGRDFRCTV GKLYPSCLEE SPWKPPTYLF STDGTEEFEL DGEMVALPPV UniProtKB: Kelch repeat and BTB domain-containing protein 2 |
-Macromolecule #3: Phosphatidylinositol 3-kinase regulatory subunit alpha
| Macromolecule | Name: Phosphatidylinositol 3-kinase regulatory subunit alpha type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.710281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAEGYQYRA LYDYKKEREE DIDLHLGDIL TVNKGSLVAL GFSDGQEARP EEIGWLNGYN ETTGERGDFP GTYVEYIGRK KISPPTPKP RPPRPLPVAP GSSKTEADVE QQALTLPDLA EQFAPPDIAP PLLIKLVEAI EKKGLECSTL YRTQSSSNLA E LRQLLDCD ...String: MSAEGYQYRA LYDYKKEREE DIDLHLGDIL TVNKGSLVAL GFSDGQEARP EEIGWLNGYN ETTGERGDFP GTYVEYIGRK KISPPTPKP RPPRPLPVAP GSSKTEADVE QQALTLPDLA EQFAPPDIAP PLLIKLVEAI EKKGLECSTL YRTQSSSNLA E LRQLLDCD TPSVDLEMID VHVLADAFKR YLLDLPNPVI PAAVYSEMIS LAPEVQSSEE YIQLLKKLIR SPSIPHQYWL TL QYLLKHF FKLSQTSSKN LLNARVLSEI FSPMLFRFSA ASSDNTENLI KVIEILISTE WNERQPAPAL PPKPPKPTTV ANN GMNNNM SLQDAEWYWG DISREEVNEK LRDTADGTFL VRDASTKMHG DYTLTLRKGG NNKLIKIFHR DGKYGFSDPL TFSS VVELI NHYRNESLAQ YNPKLDVKLL YPVSKYQQDQ VVKEDNIEAV GKKLHEYNTQ FQEKSREYDR LYEEYTRTSQ EIQMK RTAI EAFNETIKIF EEQCQTQERY SKEYIEKFKR EGNEKEIQRI MHNYDKLKSR ISEIIDSRRR LEEDLKKQAA EYREID KRM NSIKPDLIQL RKTRDQYLMW LTQKGVRQKK LNEWLGNENT EDQYSLVEDD EDLPHHDEKT WNVGSSNRNK AENLLRG KR DGTFLVRESS KQGCYACSVV VDGEVKHCVI NKTATGYGFA EPYNLYSSLK ELVLHYQHTS LVQHNDSLNV TLAYPVYA Q QRR UniProtKB: Phosphatidylinositol 3-kinase regulatory subunit alpha |
-Macromolecule #4: Cullin-3
| Macromolecule | Name: Cullin-3 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.063328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNLSKGTGS RKDTKMRIRA FPMTMDEKYV NSIWDLLKNA IQEIQRKNNS GLSFEELYRN AYTMVLHKHG EKLYTGLREV VTEHLINKV REDVLNSLNN NFLQTLNQAW NDHQTAMVMI RDILMYMDRV YVQQNNVENV YNLGLIIFRD QVVRYGCIRD H LRQTLLDM ...String: MSNLSKGTGS RKDTKMRIRA FPMTMDEKYV NSIWDLLKNA IQEIQRKNNS GLSFEELYRN AYTMVLHKHG EKLYTGLREV VTEHLINKV REDVLNSLNN NFLQTLNQAW NDHQTAMVMI RDILMYMDRV YVQQNNVENV YNLGLIIFRD QVVRYGCIRD H LRQTLLDM IARERKGEVV DRGAIRNACQ MLMILGLEGR SVYEEDFEAP FLEMSAEFFQ MESQKFLAEN SASVYIKKVE AR INEEIER VMHCLDKSTE EPIVKVVERE LISKHMKTIV EMENSGLVHM LKNGKTEDLG CMYKLFSRVP NGLKTMCECM SSY LREQGK ALVSEEGEGK NPVDYIQGLL DLKSRFDRFL LESFNNDRLF KQTIAGDFEY FLNLNSRSPE YLSLFIDDKL KKGV KGLTE QEVETILDKA MVLFRFMQEK DVFERYYKQH LARRLLTNKS VSDDSEKNMI SKLKTECGCQ FTSKLEGMFR DMSIS NTTM DEFRQHLQAT GVSLGGVDLT VRVLTTGYWP TQSATPKCNI PPAPRHAFEI FRRFYLAKHS GRQLTLQHHM GSADLN ATF YGPVKKEDGS EVGVGGAQVT GSNTRKHILQ VSTFQMTILM LFNNREKYTF EEIQQETDIP ERELVRALQS LACGKPT QR VLTKEPKSKE IENGHIFTVN DQFTSKLHRV KIQTVAAKQG ESDPERKETR QKVDDDRKHE IEAAIVRIMK SRKKMQHN V LVAEVTQQLK ARFLPSPVVI KKRIEGLIER EYLARTPEDR KVYTYVA UniProtKB: Cullin-3 |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: 7CIO |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 4.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 333476 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller
















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)