[English] 日本語
 Yorodumi
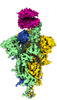
Yorodumi- EMDB-32176: SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C1 state -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32176 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C1 state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / coronavirus / Beta variant / B.1.351 lineage / spike protein / VIRAL PROTEIN / VIRAL PROTEIN-HYDROLASE complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / viral translation / regulation of systemic arterial blood pressure by renin-angiotensin / positive regulation of gap junction assembly / tryptophan transport / regulation of cardiac conduction ...positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / viral translation / regulation of systemic arterial blood pressure by renin-angiotensin / positive regulation of gap junction assembly / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy / peptidyl-dipeptidase activity / regulation of vasoconstriction / transporter activator activity / Metabolism of Angiotensinogen to Angiotensins / carboxypeptidase activity / angiotensin maturation / viral life cycle / Attachment and Entry / receptor-mediated endocytosis of virus by host cell / metallocarboxypeptidase activity / positive regulation of cardiac muscle contraction / regulation of cytokine production / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / negative regulation of ERK1 and ERK2 cascade / positive regulation of reactive oxygen species metabolic process / metallopeptidase activity / endocytic vesicle membrane / regulation of cell population proliferation / virus receptor activity / regulation of inflammatory response / endopeptidase activity / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / Potential therapeutics for SARS / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / apical plasma membrane / host cell surface receptor binding / cilium / symbiont-mediated suppression of host innate immune response / membrane raft / endocytosis involved in viral entry into host cell / endoplasmic reticulum lumen / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / cell surface / negative regulation of transcription by RNA polymerase II / extracellular space / extracellular exosome / extracellular region / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Xu C / Cong Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM. Authors: Yifan Wang / Cong Xu / Yanxing Wang / Qin Hong / Chao Zhang / Zuyang Li / Shiqi Xu / Qinyu Zuo / Caixuan Liu / Zhong Huang / Yao Cong /  Abstract: The emergence of SARS-CoV-2 Kappa and Beta variants with enhanced transmissibility and resistance to neutralizing antibodies has created new challenges for the control of the ongoing COVID-19 ...The emergence of SARS-CoV-2 Kappa and Beta variants with enhanced transmissibility and resistance to neutralizing antibodies has created new challenges for the control of the ongoing COVID-19 pandemic. Understanding the structural nature of Kappa and Beta spike (S) proteins and their association with ACE2 is of significant importance. Here we present two cryo-EM structures for each of the Kappa and Beta spikes in the open and open-prone transition states. Compared with wild-type (WT) or G614 spikes, the two variant spikes appear more untwisted/open especially for Beta, and display a considerable population shift towards the open state as well as more pronounced conformational dynamics. Moreover, we capture four conformational states of the S-trimer/ACE2 complex for each of the two variants, revealing an enlarged conformational landscape for the Kappa and Beta S-ACE2 complexes and pronounced population shift towards the three RBDs up conformation. These results implicate that the mutations in Kappa and Beta may modify the kinetics of receptor binding and viral fusion to improve virus fitness. Combined with biochemical analysis, our structural study shows that the two variants are enabled to efficiently interact with ACE2 receptor despite their sensitive ACE2 binding surface is modified to escape recognition by some potent neutralizing MAbs. Our findings shed new light on the pathogenicity and immune evasion mechanism of the Beta and Kappa variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32176.map.gz emd_32176.map.gz | 17.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32176-v30.xml emd-32176-v30.xml emd-32176.xml emd-32176.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32176.png emd_32176.png | 93.9 KB | ||
| Filedesc metadata |  emd-32176.cif.gz emd-32176.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32176 http://ftp.pdbj.org/pub/emdb/structures/EMD-32176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32176 | HTTPS FTP |
-Related structure data
| Related structure data |  7vxdMC  7vx1C  7vx4C  7vx5C  7vx9C  7vxaC  7vxbC  7vxcC  7vxeC  7vxfC  7vxiC  7vxkC  7vxmC  7wevC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32176.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32176.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.093 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 spike protein in complex with ACE2, Beta Variant
| Entire | Name: SARS-CoV-2 spike protein in complex with ACE2, Beta Variant |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike protein in complex with ACE2, Beta Variant
| Supramolecule | Name: SARS-CoV-2 spike protein in complex with ACE2, Beta Variant type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 spike protein
| Supramolecule | Name: SARS-CoV-2 spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: ACE2
| Supramolecule | Name: ACE2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 139.650516 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNFTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFA NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNFTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFA NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRGLPQ GFSALEPLVD LPIGINITRF QT LHISYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNITNLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIR GDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGV KGFN CYFPLQSYGF QPTYGVGYQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLT GTGVLTESNK KFLPFQ QFG RDIADTTDAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQGVN CTEVPVAIHA DQLTPTWRVY STGSNVF QT RAGCLIGAEH VNNSYECDIP IGAGICASYQ TQTNSPGSAS SVASQSIIAY TMSLGVENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLN RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KDFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFNGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGA ALQIPFAMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTA SALGKLQDVV NQNAQALNTL V KQLSSNFG AISSVLNDIL SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ GSGYIPEAPR DGQAYVRKDG EWVLLSTFLE NLYFQGDYKD DDDKHHHHHH HHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: Angiotensin-converting enzyme 2
| Macromolecule | Name: Angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: angiotensin-converting enzyme 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.396492 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHSSALLCCL VLLTGVRAQS TIEEQAKTFL DKFNHEAEDL FYQSSLASWN YNTNITEENV QNMNNAGDKW SAFLKEQSTL AQMYPLQEI QNLTVKLQLQ ALQQNGSSVL SEDKSKRLNT ILNTMSTIYS TGKVCNPDNP QECLLLEPGL NEIMANSLDY N ERLWAWES ...String: MHSSALLCCL VLLTGVRAQS TIEEQAKTFL DKFNHEAEDL FYQSSLASWN YNTNITEENV QNMNNAGDKW SAFLKEQSTL AQMYPLQEI QNLTVKLQLQ ALQQNGSSVL SEDKSKRLNT ILNTMSTIYS TGKVCNPDNP QECLLLEPGL NEIMANSLDY N ERLWAWES WRSEVGKQLR PLYEEYVVLK NEMARANHYE DYGDYWRGDY EVNGVDGYDY SRGQLIEDVE HTFEEIKPLY EH LHAYVRA KLMNAYPSYI SPIGCLPAHL LGDMWGRFWT NLYSLTVPFG QKPNIDVTDA MVDQAWDAQR IFKEAEKFFV SVG LPNMTQ GFWENSMLTD PGNVQKAVCH PTAWDLGKGD FRILMCTKVT MDDFLTAHHE MGHIQYDMAY AAQPFLLRNG ANEG FHEAV GEIMSLSAAT PKHLKSIGLL SPDFQEDNET EINFLLKQAL TIVGTLPFTY MLEKWRWMVF KGEIPKDQWM KKWWE MKRE IVGVVEPVPH DETYCDPASL FHVSNDYSFI RYYTRTLYQF QFQEALCQAA KHEGPLHKCD ISNSTEAGQK LFNMLR LGK SEPWTLALEN VVGAKNMNVR PLLNYFEPLF TWLKDQNKNS FVGWSTDWSP YADHHHHHHH HH UniProtKB: Angiotensin-converting enzyme 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















