[English] 日本語
 Yorodumi
Yorodumi- EMDB-32092: Cryo-EM structure of amyloid fibril formed by FUS low complexity ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32092 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of amyloid fibril formed by FUS low complexity domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | amyloid fibril / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembraneless organelle assembly / mRNA stabilization / intracellular membraneless organelle / regulation of RNA splicing / Processing of Capped Intron-Containing Pre-mRNA / postsynaptic cytosol / positive regulation of double-strand break repair via homologous recombination / presynaptic cytosol / mRNA Splicing - Major Pathway / RNA splicing ...membraneless organelle assembly / mRNA stabilization / intracellular membraneless organelle / regulation of RNA splicing / Processing of Capped Intron-Containing Pre-mRNA / postsynaptic cytosol / positive regulation of double-strand break repair via homologous recombination / presynaptic cytosol / mRNA Splicing - Major Pathway / RNA splicing / mRNA 3'-UTR binding / transcription coregulator activity / molecular condensate scaffold activity / protein homooligomerization / GABA-ergic synapse / amyloid fibril formation / transcription coactivator activity / chromatin binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / glutamatergic synapse / DNA binding / RNA binding / zinc ion binding / nucleoplasm / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Sun YP / Xia WC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: iScience / Year: 2022 Journal: iScience / Year: 2022Title: Molecular structure of an amyloid fibril formed by FUS low-complexity domain. Authors: Yunpeng Sun / Shenqing Zhang / Jiaojiao Hu / Youqi Tao / Wencheng Xia / Jinge Gu / Yichen Li / Qin Cao / Dan Li / Cong Liu /  Abstract: FUS is a multifunctional nuclear protein which undergoes liquid-liquid phase separation in response to stress and DNA damage. Dysregulation of FUS dynamic phase separation leads to formation of ...FUS is a multifunctional nuclear protein which undergoes liquid-liquid phase separation in response to stress and DNA damage. Dysregulation of FUS dynamic phase separation leads to formation of pathological fibril closely associated with neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal dementia. In this study, we determined the cryo-EM structure of a cytotoxic fibril formed by the low-complexity (LC) domain of FUS at 2.9 Å resolution. The fibril structure exhibits a new and extensive serpentine fold consisting of three motifs incorporating together via a Tyr triad. FUS LC employs 91 residues to form an enlarged and stable fibril core via hydrophilic interaction and hydrogen bonds, which is distinct from most of previously determined fibrils commonly stabilized by hydrophobic interaction. Our work reveals the structural basis underlying formation of a cytotoxic and thermostable fibril of FUS LC and sheds light on understanding the liquid-to-solid phase transition of FUS in disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32092.map.gz emd_32092.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32092-v30.xml emd-32092-v30.xml emd-32092.xml emd-32092.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32092_fsc.xml emd_32092_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32092.png emd_32092.png | 33.1 KB | ||
| Filedesc metadata |  emd-32092.cif.gz emd-32092.cif.gz | 5.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32092 http://ftp.pdbj.org/pub/emdb/structures/EMD-32092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32092 | HTTPS FTP |
-Validation report
| Summary document |  emd_32092_validation.pdf.gz emd_32092_validation.pdf.gz | 594.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32092_full_validation.pdf.gz emd_32092_full_validation.pdf.gz | 594.4 KB | Display | |
| Data in XML |  emd_32092_validation.xml.gz emd_32092_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_32092_validation.cif.gz emd_32092_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32092 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32092 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32092 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32092 | HTTPS FTP |
-Related structure data
| Related structure data |  7vqqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32092.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32092.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Amyloid fibril formed by FUS low complexity domain
| Entire | Name: Amyloid fibril formed by FUS low complexity domain |
|---|---|
| Components |
|
-Supramolecule #1: Amyloid fibril formed by FUS low complexity domain
| Supramolecule | Name: Amyloid fibril formed by FUS low complexity domain / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: fusion protein of mCerulean and FUS LCD
| Macromolecule | Name: fusion protein of mCerulean and FUS LCD / type: protein_or_peptide / ID: 1 Details: 6His-tagged mCerulean:(FPbase ID: J2JWA, Link: https://www.fpbase.org/protein/mcerulean/) Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.948352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMVSKG EELFTGVVPI LVELDGDVNG HKFSVSGEGE GDATYGKLTL KFICTTGKLP VPWPTLVTT LTWGVQCFAR YPDHMKQHDF FKSAMPEGYV QERTIFFKDD GNYKTRAEVK FEGDTLVNRI ELKGIDFKED G NILGHKLE ...String: MSYYHHHHHH DYDIPTTENL YFQGAMVSKG EELFTGVVPI LVELDGDVNG HKFSVSGEGE GDATYGKLTL KFICTTGKLP VPWPTLVTT LTWGVQCFAR YPDHMKQHDF FKSAMPEGYV QERTIFFKDD GNYKTRAEVK FEGDTLVNRI ELKGIDFKED G NILGHKLE YNAISDNVYI TADKQKNGIK ANFKIRHNIE DGSVQLADHY QQNTPIGDGP VLLPDNHYLS TQSKLSKDPN EK RDHMVLL EFVTAAGITL GMDELYKAGT MDPASNDYTQ QATQSYGAYP TQPGQGYSQQ SSQPYGQQSY SGYSQSTDTS GYG QSSYSS YGQSQNTGYG TQSTPQGYGS TGGYGSSQSS QSSYGQQSSY PGYGQQPAPS STSGSYGSSS QSSSYGQPQS GSYS QQPSY GGQQQSYGQQ QSYNPPQGYG QQNQYNSSSG GGGGGGGGGN YGQDQSSMSS GGGSGGGYGN QDQSGGGGSG GYGQQ DRG UniProtKB: RNA-binding protein FUS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


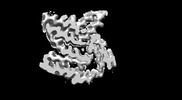













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















