+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Telomeric trinucleosome | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Telomere / nucleosome / chromatin / DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA complex | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報negative regulation of megakaryocyte differentiation / protein localization to CENP-A containing chromatin / Chromatin modifying enzymes / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / Recognition and association of DNA glycosylase with site containing an affected pyrimidine ...negative regulation of megakaryocyte differentiation / protein localization to CENP-A containing chromatin / Chromatin modifying enzymes / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / telomere organization / Interleukin-7 signaling / Inhibition of DNA recombination at telomere / RNA Polymerase I Promoter Opening / Meiotic synapsis / Assembly of the ORC complex at the origin of replication / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / SUMOylation of chromatin organization proteins / DNA methylation / Condensation of Prophase Chromosomes / epigenetic regulation of gene expression / Chromatin modifications during the maternal to zygotic transition (MZT) / SIRT1 negatively regulates rRNA expression / HCMV Late Events / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / innate immune response in mucosa / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / HDMs demethylate histones / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / HDACs deacetylate histones / Nonhomologous End-Joining (NHEJ) / RNA Polymerase I Promoter Escape / Transcriptional regulation by small RNAs / Formation of the beta-catenin:TCF transactivating complex / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / G2/M DNA damage checkpoint / Metalloprotease DUBs / NoRC negatively regulates rRNA expression / DNA Damage/Telomere Stress Induced Senescence / B-WICH complex positively regulates rRNA expression / PKMTs methylate histone lysines / Meiotic recombination / Pre-NOTCH Transcription and Translation / RMTs methylate histone arginines / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Transcriptional regulation of granulopoiesis / UCH proteinases / HCMV Early Events / antimicrobial humoral immune response mediated by antimicrobial peptide / structural constituent of chromatin / antibacterial humoral response / E3 ubiquitin ligases ubiquitinate target proteins / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / nucleosome / heterochromatin formation / RUNX1 regulates transcription of genes involved in differentiation of HSCs / nucleosome assembly / Processing of DNA double-strand break ends / HATs acetylate histones / Senescence-Associated Secretory Phenotype (SASP) / Factors involved in megakaryocyte development and platelet production / chromatin organization / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / Oxidative Stress Induced Senescence / defense response to Gram-negative bacterium / gene expression / Estrogen-dependent gene expression / killing of cells of another organism / chromosome, telomeric region / Ub-specific processing proteases / defense response to Gram-positive bacterium / cadherin binding / Amyloid fiber formation / protein heterodimerization activity / negative regulation of cell population proliferation / protein-containing complex / extracellular space / DNA binding / RNA binding / extracellular exosome / extracellular region / nucleoplasm / nucleus / membrane / cytosol 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
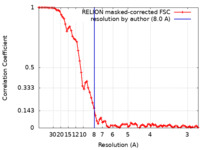
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 8.0 Å | |||||||||
 データ登録者 データ登録者 | Soman A | |||||||||
| 資金援助 |  シンガポール, 1件 シンガポール, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2022 ジャーナル: Nature / 年: 2022タイトル: Columnar structure of human telomeric chromatin. 著者: Aghil Soman / Sook Yi Wong / Nikolay Korolev / Wahyu Surya / Simon Lattmann / Vinod K Vogirala / Qinming Chen / Nikolay V Berezhnoy / John van Noort / Daniela Rhodes / Lars Nordenskiöld /    要旨: Telomeres, the ends of eukaryotic chromosomes, play pivotal parts in ageing and cancer and are targets of DNA damage and the DNA damage response. Little is known about the structure of telomeric ...Telomeres, the ends of eukaryotic chromosomes, play pivotal parts in ageing and cancer and are targets of DNA damage and the DNA damage response. Little is known about the structure of telomeric chromatin at the molecular level. Here we used negative stain electron microscopy and single-molecule magnetic tweezers to characterize 3-kbp-long telomeric chromatin fibres. We also obtained the cryogenic electron microscopy structure of the condensed telomeric tetranucleosome and its dinucleosome unit. The structure displayed close stacking of nucleosomes with a columnar arrangement, and an unusually short nucleosome repeat length that comprised about 132 bp DNA wound in a continuous superhelix around histone octamers. This columnar structure is primarily stabilized by the H2A carboxy-terminal and histone amino-terminal tails in a synergistic manner. The columnar conformation results in exposure of the DNA helix, which may make it susceptible to both DNA damage and the DNA damage response. The conformation also exists in an alternative open state, in which one nucleosome is unstacked and flipped out, which exposes the acidic patch of the histone surface. The structural features revealed in this work suggest mechanisms by which protein factors involved in telomere maintenance can access telomeric chromatin in its compact form. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_31816.map.gz emd_31816.map.gz | 38.7 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-31816-v30.xml emd-31816-v30.xml emd-31816.xml emd-31816.xml | 22.8 KB 22.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_31816_fsc.xml emd_31816_fsc.xml | 8.1 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_31816.png emd_31816.png | 60.4 KB | ||
| マスクデータ |  emd_31816_msk_1.map emd_31816_msk_1.map | 42.9 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-31816.cif.gz emd-31816.cif.gz | 6 KB | ||
| その他 |  emd_31816_half_map_1.map.gz emd_31816_half_map_1.map.gz emd_31816_half_map_2.map.gz emd_31816_half_map_2.map.gz | 33.1 MB 33 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31816 http://ftp.pdbj.org/pub/emdb/structures/EMD-31816 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31816 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31816 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_31816_validation.pdf.gz emd_31816_validation.pdf.gz | 971 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_31816_full_validation.pdf.gz emd_31816_full_validation.pdf.gz | 970.6 KB | 表示 | |
| XML形式データ |  emd_31816_validation.xml.gz emd_31816_validation.xml.gz | 13.6 KB | 表示 | |
| CIF形式データ |  emd_31816_validation.cif.gz emd_31816_validation.cif.gz | 19.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31816 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31816 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31816 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31816 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7v9jMC  7v90C  7v96C  7v9cC  7v9kC  7v9sC  7va4C M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_31816.map.gz / 形式: CCP4 / 大きさ: 42.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_31816.map.gz / 形式: CCP4 / 大きさ: 42.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_31816_msk_1.map emd_31816_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
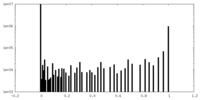
| 投影像・断面図 |
| ||||||||||||
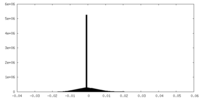
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_31816_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
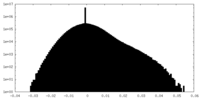
| 投影像・断面図 |
| ||||||||||||
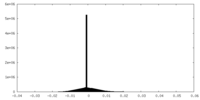
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_31816_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Telomeric Mononucleosome
| 全体 | 名称: Telomeric Mononucleosome |
|---|---|
| 要素 |
|
-超分子 #1: Telomeric Mononucleosome
| 超分子 | 名称: Telomeric Mononucleosome / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all 詳細: Mononucleosome in open statae in a telomeric tetranucleosome |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Histone H3.1
| 分子 | 名称: Histone H3.1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 6 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 15.437167 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VMALQEACEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.1 |
-分子 #2: Histone H4
| 分子 | 名称: Histone H4 / タイプ: protein_or_peptide / ID: 2 / コピー数: 6 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 11.394426 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-分子 #3: Histone H2A type 1-B/E
| 分子 | 名称: Histone H2A type 1-B/E / タイプ: protein_or_peptide / ID: 3 / コピー数: 6 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 14.165551 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSGRGKQGGK ARAKAKTRSS RAGLQFPVGR VHRLLRKGNY SERVGAGAPV YLAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAIR NDEELNKLLG RVTIAQGGVL PNIQAVLLPK KTESHHKAKG K UniProtKB: Histone H2A type 1-B/E |
-分子 #4: Histone H2B type 1-K
| 分子 | 名称: Histone H2B type 1-K / タイプ: protein_or_peptide / ID: 4 / コピー数: 6 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 11.151906 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: KKRKRSRKES YSVYVYKVLK QVHPDTGISS KAMGIMNSFV NDIFERIAGE ASRLAHYNKR STITSREIQT AVRLLLPGEL AKHAVSEGT KAVTKYTSAK UniProtKB: Histone H2B type 1-K |
-分子 #5: DNA (408-mer)
| 分子 | 名称: DNA (408-mer) / タイプ: dna / ID: 5 / コピー数: 1 / 分類: DNA |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 129.781156 KDa |
| 配列 | 文字列: (DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG) (DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA) (DG) (DG)(DG)(DT)(DT)(DA) ...文字列: (DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG) (DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA) (DG) (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG) (DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG) (DT)(DT)(DA)(DG)(DG)(DG)(DT) (DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG) (DG)(DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG) (DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA) (DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT) (DT)(DA)(DG)(DG)(DG) (DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG) (DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA) (DG)(DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG) (DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG) (DT) (DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG) (DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG) (DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT) (DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG) (DG) (DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG) (DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT)(DA) (DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT) (DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG) (DT)(DT)(DA)(DG)(DG) (DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA) (DG)(DG)(DG)(DT)(DT) (DA)(DG)(DG)(DG) (DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG)(DT) (DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG) (DG)(DT)(DT)(DA)(DG)(DG)(DG) (DT)(DT) (DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG) (DT)(DT)(DA)(DG)(DG)(DG)(DT)(DT) (DA) (DG)(DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG)(DT) (DT)(DA)(DG)(DG)(DG)(DT)(DT)(DA)(DG) (DG)(DG)(DT)(DT)(DA)(DG)(DG)(DG) |
-分子 #6: DNA (408-mer)
| 分子 | 名称: DNA (408-mer) / タイプ: dna / ID: 6 / コピー数: 1 / 分類: DNA |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 122.229008 KDa |
| 配列 | 文字列: (DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA) (DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC) (DT) (DA)(DA)(DC)(DC)(DC) ...文字列: (DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA) (DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC) (DT) (DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA) (DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA) (DC)(DC)(DC)(DT)(DA)(DA)(DC) (DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA) (DA)(DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA) (DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC) (DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC) (DC)(DC)(DT)(DA)(DA) (DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA) (DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC) (DT)(DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA) (DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA) (DC) (DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA) (DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA) (DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC) (DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA) (DA) (DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA) (DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC)(DC) (DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC) (DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA)(DA) (DC)(DC)(DC)(DT)(DA) (DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC) (DT)(DA)(DA)(DC)(DC) (DC)(DT)(DA)(DA) (DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA)(DC) (DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA) (DA)(DC)(DC)(DC)(DT)(DA)(DA) (DC)(DC) (DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA) (DC)(DC)(DC)(DT)(DA)(DA)(DC)(DC) (DC) (DT)(DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA)(DC) (DC)(DC)(DT)(DA)(DA)(DC)(DC)(DC)(DT) (DA)(DA)(DC)(DC)(DC)(DT)(DA)(DA) |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 1 mg/mL |
|---|---|
| 緩衝液 | pH: 6 |
| グリッド | モデル: Quantifoil R1.2/1.3 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % |
| 詳細 | tri-NCP subunit of telomeric tetranucleosome |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: OTHER / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)