+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Telomeric Dinucleosome at 5angstroms | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.0 Å | |||||||||
 Authors Authors | Soman A | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Columnar structure of human telomeric chromatin. Authors: Aghil Soman / Sook Yi Wong / Nikolay Korolev / Wahyu Surya / Simon Lattmann / Vinod K Vogirala / Qinming Chen / Nikolay V Berezhnoy / John van Noort / Daniela Rhodes / Lars Nordenskiöld /    Abstract: Telomeres, the ends of eukaryotic chromosomes, play pivotal parts in ageing and cancer and are targets of DNA damage and the DNA damage response. Little is known about the structure of telomeric ...Telomeres, the ends of eukaryotic chromosomes, play pivotal parts in ageing and cancer and are targets of DNA damage and the DNA damage response. Little is known about the structure of telomeric chromatin at the molecular level. Here we used negative stain electron microscopy and single-molecule magnetic tweezers to characterize 3-kbp-long telomeric chromatin fibres. We also obtained the cryogenic electron microscopy structure of the condensed telomeric tetranucleosome and its dinucleosome unit. The structure displayed close stacking of nucleosomes with a columnar arrangement, and an unusually short nucleosome repeat length that comprised about 132 bp DNA wound in a continuous superhelix around histone octamers. This columnar structure is primarily stabilized by the H2A carboxy-terminal and histone amino-terminal tails in a synergistic manner. The columnar conformation results in exposure of the DNA helix, which may make it susceptible to both DNA damage and the DNA damage response. The conformation also exists in an alternative open state, in which one nucleosome is unstacked and flipped out, which exposes the acidic patch of the histone surface. The structural features revealed in this work suggest mechanisms by which protein factors involved in telomere maintenance can access telomeric chromatin in its compact form. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31908.map.gz emd_31908.map.gz | 14.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31908-v30.xml emd-31908-v30.xml emd-31908.xml emd-31908.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
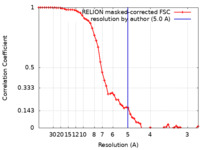
| FSC (resolution estimation) |  emd_31908_fsc.xml emd_31908_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_31908.png emd_31908.png | 53.2 KB | ||
| Masks |  emd_31908_msk_1.map emd_31908_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Others |  emd_31908_half_map_1.map.gz emd_31908_half_map_1.map.gz emd_31908_half_map_2.map.gz emd_31908_half_map_2.map.gz | 11.1 MB 11.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31908 http://ftp.pdbj.org/pub/emdb/structures/EMD-31908 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31908 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31908 | HTTPS FTP |
-Validation report
| Summary document |  emd_31908_validation.pdf.gz emd_31908_validation.pdf.gz | 754.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31908_full_validation.pdf.gz emd_31908_full_validation.pdf.gz | 754 KB | Display | |
| Data in XML |  emd_31908_validation.xml.gz emd_31908_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_31908_validation.cif.gz emd_31908_validation.cif.gz | 15.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31908 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31908 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31908 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31908 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31908.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31908.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31908_msk_1.map emd_31908_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_31908_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_31908_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Telomeric Dinucleosome
| Entire | Name: Telomeric Dinucleosome |
|---|---|
| Components |
|
-Supramolecule #1: Telomeric Dinucleosome
| Supramolecule | Name: Telomeric Dinucleosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)