+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of an alphavirus, Getah virus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Virus / mature / Infective | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=4 icosahedral viral capsid / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / membrane Similarity search - Function | |||||||||
| Biological species |  Getah virus Getah virus | |||||||||
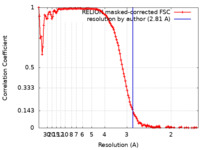
| Method | single particle reconstruction / cryo EM / Resolution: 2.81 Å | |||||||||
 Authors Authors | Liu Z / Liu C / Wang A | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2022 Journal: Cell Discov / Year: 2022Title: Structure of infective Getah virus at 2.8 Å resolution determined by cryo-electron microscopy. Authors: Aojie Wang / Feng Zhou / Congcong Liu / Dongsheng Gao / Ruxi Qi / Yiheng Yin / Sheng Liu / Yuanzhu Gao / Lutang Fu / Yinhe Xia / Yawei Xu / Chuanqing Wang / Zheng Liu /  Abstract: Getah virus (GETV), a member of the genus alphavirus, is a mosquito-borne pathogen that can cause pyrexia and reproductive losses in animals. Although antibodies to GETV have been found in over 10% ...Getah virus (GETV), a member of the genus alphavirus, is a mosquito-borne pathogen that can cause pyrexia and reproductive losses in animals. Although antibodies to GETV have been found in over 10% of healthy people, there are no reports of clinical symptoms associated with GETV. The biological and pathological properties of GETV are largely unknown and antiviral or vaccine treatments against GETV are still unavailable due to a lack of knowledge of the structure of the GETV virion. Here, we present the structure of infective GETV at a resolution of 2.8 Å with the atomic models of the capsid protein and the envelope glycoproteins E1 and E2. We have identified numerous glycosylation and S-acylation sites in E1 and E2. The surface-exposed glycans indicate a possible impact on viral immune evasion and host cell invasion. The S-acylation sites might be involved in stabilizing the transmembrane assembly of E1 and E2. In addition, a cholesterol and a phospholipid molecule are observed in a transmembrane hydrophobic pocket, together with two more cholesterols surrounding the pocket. The cholesterol and phospholipid stabilize the hydrophobic pocket in the viral envelope membrane. The structural information will assist structure-based antiviral and vaccine screening, design, and optimization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31533.map.gz emd_31533.map.gz | 452.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31533-v30.xml emd-31533-v30.xml emd-31533.xml emd-31533.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31533_fsc.xml emd_31533_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_31533.png emd_31533.png | 107.1 KB | ||
| Filedesc metadata |  emd-31533.cif.gz emd-31533.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31533 http://ftp.pdbj.org/pub/emdb/structures/EMD-31533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31533 | HTTPS FTP |
-Related structure data
| Related structure data |  7fd2MC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31533.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31533.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.66 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Getah virus
| Entire | Name:  Getah virus Getah virus |
|---|---|
| Components |
|
-Supramolecule #1: Getah virus
| Supramolecule | Name: Getah virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / NCBI-ID: 59300 / Sci species name: Getah virus / Sci species strain: GETV-V1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Envelope glycoprotein 1
| Macromolecule | Name: Envelope glycoprotein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:  Getah virus Getah virus |
| Molecular weight | Theoretical: 47.560648 KDa |
| Sequence | String: YEHTATIPNV VGFPYKAHIE RNGFSPMTLQ LEVLGTSLEP TLNLEYITCE YKTVVPSPYI KCCGTSECRS MERPDYQCQV YTGVYPFMW GGAYCFCDTE NTQLSEAYVD RSDVCKHDHA AAYKAHTAAM KATIRISYGN LNQTTTAFVN GEHTVTVGGS R FTFGPIST ...String: YEHTATIPNV VGFPYKAHIE RNGFSPMTLQ LEVLGTSLEP TLNLEYITCE YKTVVPSPYI KCCGTSECRS MERPDYQCQV YTGVYPFMW GGAYCFCDTE NTQLSEAYVD RSDVCKHDHA AAYKAHTAAM KATIRISYGN LNQTTTAFVN GEHTVTVGGS R FTFGPIST AWTPFDNKIV VYKNDVYNQD FPPYGSGQPG RFGDIQSRTV ESKDLYANTA LKLSRPSSGT VHVPYTQTPS GF KYWLKER GTSLNDKAPF GCVIKTNPVR AENCAVGNIP VSMDIPDTAF TRVIDAPAVT NLECQVAVCT HSSDFGGIAT LTF KTDKPG KCAVHSHSNV ATIQEAAVDI KTDGKITLHF STASASPAFK VSVCSAKTTC TAACEPPKDH IVPYGASHNN QVFP DMSGT AMTWVQRVAG GLGGLTLAAV AVLILVTCVT MRR UniProtKB: Structural polyprotein |
-Macromolecule #2: Envelope glycoprotein 2
| Macromolecule | Name: Envelope glycoprotein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:  Getah virus Getah virus |
| Molecular weight | Theoretical: 46.147492 KDa |
| Sequence | String: SVTEHFNVYK ATKPYLAYCA DCGDGQFCYS PVAIEKIRDE ASDGMIKIQV AAQIGINKGG THEHNKIRYI AGHDMKEANR DSLQVYTSG VCAIRGTMGH FIVAYCPPGD ELKVQFQDAE SHTQACKVQY KHAPAPVGRE KFTVRPHFGI EVPCTTYQLT T APTEEEID ...String: SVTEHFNVYK ATKPYLAYCA DCGDGQFCYS PVAIEKIRDE ASDGMIKIQV AAQIGINKGG THEHNKIRYI AGHDMKEANR DSLQVYTSG VCAIRGTMGH FIVAYCPPGD ELKVQFQDAE SHTQACKVQY KHAPAPVGRE KFTVRPHFGI EVPCTTYQLT T APTEEEID MHTPPDIPDI TLLSQQSGNV KITAGGKTIR YNCTCGSGNV GTTSSDKTIN SCKIAQCHAA VTNHDKWQYT SS FVPRADQ LSRKGKVHVP FPLTNSTCRV PVARAPGVTY GKRELTVKLH PDHPTLLTYR SLGADPRPYE EWIDRYVERT IPV TEDGIE YRWGNNPPVR LWAQLTTEGK PHGWPHEIIL YYYGLYPAAT IAAVSAAGLA VVLSLLASCY MFATARRKCL TPYA LTPGA VVPVTLGVLC CAPR UniProtKB: Structural polyprotein |
-Macromolecule #3: capsid protein
| Macromolecule | Name: capsid protein / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:  Getah virus Getah virus |
| Molecular weight | Theoretical: 30.200117 KDa |
| Sequence | String: MNYIPTQTFY GRRWRPRPAY RPWRVPMQPA PPMVIPELQT PIVQAQQMQQ LISAVSALTT KQNGKAPKKP KKKPQKAKAK KNEQQKKNE NKKPPPKQKN PAKKKKPGKR ERMCMKIEND CIFEVKLDGK VTGYACLVGD KVMKPAHVKG VIDNPDLAKL T YKKSSKYD ...String: MNYIPTQTFY GRRWRPRPAY RPWRVPMQPA PPMVIPELQT PIVQAQQMQQ LISAVSALTT KQNGKAPKKP KKKPQKAKAK KNEQQKKNE NKKPPPKQKN PAKKKKPGKR ERMCMKIEND CIFEVKLDGK VTGYACLVGD KVMKPAHVKG VIDNPDLAKL T YKKSSKYD LECAQIPVHM KSDASKYTHE KPEGHYNWHH GAVQYSGGRF TIPTGAGKPG DSGRPIFDNK GRVVAIVLGG AN EGARTAL SVVTWTKDMV TRYTPEGTEE W UniProtKB: Structural polyprotein |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 8 / Number of copies: 4 / Formula: PCW |
|---|---|
| Molecular weight | Theoretical: 787.121 Da |
| Chemical component information |  ChemComp-PCW: |
-Macromolecule #9: STEARIC ACID
| Macromolecule | Name: STEARIC ACID / type: ligand / ID: 9 / Number of copies: 8 / Formula: STE |
|---|---|
| Molecular weight | Theoretical: 284.477 Da |
| Chemical component information |  ChemComp-STE: |
-Macromolecule #10: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 10 / Number of copies: 12 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #11: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 11 / Number of copies: 12 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: NICKEL/TITANIUM / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.00038 kPa |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 16894 / Average electron dose: 1.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||
| Output model |  PDB-7fd2: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)