[English] 日本語
 Yorodumi
Yorodumi- EMDB-31422: Cryo-EM structure of the chemokine receptor CCR5 in complex with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31422 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the chemokine receptor CCR5 in complex with MIP-1a and Gi | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G Protein-coupled receptor / Chemokine Receptor CCR5 / MIP-1a / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationchemokine (C-C motif) ligand 5 binding / lymphocyte chemotaxis / granulocyte chemotaxis / CCR1 chemokine receptor binding / positive regulation of microglial cell migration / positive regulation of natural killer cell chemotaxis / negative regulation of macrophage apoptotic process / regulation of behavior / astrocyte cell migration / chemokine receptor activity ...chemokine (C-C motif) ligand 5 binding / lymphocyte chemotaxis / granulocyte chemotaxis / CCR1 chemokine receptor binding / positive regulation of microglial cell migration / positive regulation of natural killer cell chemotaxis / negative regulation of macrophage apoptotic process / regulation of behavior / astrocyte cell migration / chemokine receptor activity / CCR5 chemokine receptor binding / eosinophil degranulation / CCR chemokine receptor binding / regulation of sensory perception of pain / signaling / negative regulation of bone mineralization / positive regulation of microglial cell activation / cell activation / C-C chemokine receptor activity / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / T cell chemotaxis / chemokine-mediated signaling pathway / C-C chemokine binding / eosinophil chemotaxis / positive regulation of calcium ion transport / response to cholesterol / chemokine activity / Chemokine receptors bind chemokines / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / dendritic cell chemotaxis / phospholipase activator activity / Interleukin-10 signaling / positive regulation of calcium ion import / chemoattractant activity / macrophage chemotaxis / exocytosis / negative regulation of osteoclast differentiation / monocyte chemotaxis / cellular response to interleukin-1 / negative regulation by host of viral transcription / Binding and entry of HIV virion / cellular defense response / positive regulation of protein localization to cell cortex / Adenylate cyclase inhibitory pathway / T cell migration / coreceptor activity / D2 dopamine receptor binding / response to prostaglandin E / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cytoskeleton organization / neutrophil chemotaxis / cellular response to forskolin / regulation of mitotic spindle organization / positive regulation of calcium-mediated signaling / positive regulation of interleukin-1 beta production / cell chemotaxis / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / G protein-coupled receptor binding / calcium-mediated signaling / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / cellular response to type II interferon / response to peptide hormone / G-protein beta/gamma-subunit complex binding / centriolar satellite / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / response to toxic substance / G-protein activation / intracellular calcium ion homeostasis / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / positive regulation of inflammatory response / Prostacyclin signalling through prostacyclin receptor / antimicrobial humoral immune response mediated by antimicrobial peptide / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / chemotaxis / photoreceptor disc membrane / calcium ion transport / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / positive regulation of tumor necrosis factor production / osteoblast differentiation / Vasopressin regulates renal water homeostasis via Aquaporins / GDP binding / kinase activity / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Zhang H / Chen K | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5. Authors: Hui Zhang / Kun Chen / Qiuxiang Tan / Qiang Shao / Shuo Han / Chenhui Zhang / Cuiying Yi / Xiaojing Chu / Ya Zhu / Yechun Xu / Qiang Zhao / Beili Wu /  Abstract: The chemokine receptor CCR5 plays a vital role in immune surveillance and inflammation. However, molecular details that govern its endogenous chemokine recognition and receptor activation remain ...The chemokine receptor CCR5 plays a vital role in immune surveillance and inflammation. However, molecular details that govern its endogenous chemokine recognition and receptor activation remain elusive. Here we report three cryo-electron microscopy structures of G protein-coupled CCR5 in a ligand-free state and in complex with the chemokine MIP-1α or RANTES, as well as the crystal structure of MIP-1α-bound CCR5. These structures reveal distinct binding modes of the two chemokines and a specific accommodate pattern of the chemokine for the distal N terminus of CCR5. Together with functional data, the structures demonstrate that chemokine-induced rearrangement of toggle switch and plasticity of the receptor extracellular region are critical for receptor activation, while a conserved tryptophan residue in helix II acts as a trigger of receptor constitutive activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
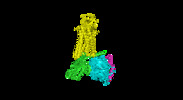
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31422.map.gz emd_31422.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31422-v30.xml emd-31422-v30.xml emd-31422.xml emd-31422.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31422.png emd_31422.png | 26 KB | ||
| Filedesc metadata |  emd-31422.cif.gz emd-31422.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31422 http://ftp.pdbj.org/pub/emdb/structures/EMD-31422 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31422 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31422 | HTTPS FTP |
-Related structure data
| Related structure data |  7f1qMC  7f1rC  7f1sC  7f1tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31422.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31422.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Chemokine receptor CCR5 in complex with MIP-1a and Gi
| Entire | Name: Chemokine receptor CCR5 in complex with MIP-1a and Gi |
|---|---|
| Components |
|
-Supramolecule #1: Chemokine receptor CCR5 in complex with MIP-1a and Gi
| Supramolecule | Name: Chemokine receptor CCR5 in complex with MIP-1a and Gi / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: C-C motif chemokine 3,C-C chemokine receptor type 5
| Macromolecule | Name: C-C motif chemokine 3,C-C chemokine receptor type 5 / type: protein_or_peptide / ID: 1 / Details: Fusion protein of MIP-1a and CCR5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.241047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SLAADTPTAC CFSYCSRQIP QNFIADYFET SSQCSKPGVI FLTKRSRQVC ADPSEEWVQK YVSDLELSAG SGSGSGSGSG SGSGSGSGS GSGSDYQVSS PIYDINYYCS EPCQKINVKQ IAARLLPPLY SLVFIFGFVG NMLVILILIN CKRLKSMTDI Y LLNLAISD ...String: SLAADTPTAC CFSYCSRQIP QNFIADYFET SSQCSKPGVI FLTKRSRQVC ADPSEEWVQK YVSDLELSAG SGSGSGSGSG SGSGSGSGS GSGSDYQVSS PIYDINYYCS EPCQKINVKQ IAARLLPPLY SLVFIFGFVG NMLVILILIN CKRLKSMTDI Y LLNLAISD LFFLLTVPFW AHYAAAQWDF GNTMCQLLTG LYFIGFFSGI FFIILLTIDR YLAVVHAVFA LKARTVTFGV VT SVITWVV AVFASLPNII FTRSQKEGLH YTCSSHFPYS QYQFWKNFQT LKIVILGLVL PLLVMVICYS GILKTLLRCR NEK KRHRAV RLIFTIMIVY FLFWAPYNIV LLLNTFQEFF GLNNCSSSNR LDQAMQVTET LGMTHCCINP IIYAFVGEKF RNYL LVFFQ KHIAKRLEVL FQGPGSWSHP QFEKGSGAGA SAGSWSHPQF EKGSDYKDDD DK UniProtKB: C-C motif chemokine 3, C-C chemokine receptor type 5 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.447141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKCTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKCTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVTAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 2.1875 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 7095732 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)