+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31137 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
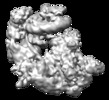
| Title | The structure of SWI/SNF-nucleosome complex | |||||||||
 Map data Map data | The structure of SWI/SNF-nucleosome complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Chromatin remodeler complex / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcarbon catabolite activation of transcription / RHO GTPases activate IQGAPs / RHO GTPases Activate WASPs and WAVEs / Regulation of actin dynamics for phagocytic cup formation / positive regulation of cell adhesion involved in single-species biofilm formation / positive regulation of mating type switching / positive regulation of invasive growth in response to glucose limitation / aggrephagy / Platelet degranulation / HDACs deacetylate histones ...carbon catabolite activation of transcription / RHO GTPases activate IQGAPs / RHO GTPases Activate WASPs and WAVEs / Regulation of actin dynamics for phagocytic cup formation / positive regulation of cell adhesion involved in single-species biofilm formation / positive regulation of mating type switching / positive regulation of invasive growth in response to glucose limitation / aggrephagy / Platelet degranulation / HDACs deacetylate histones / DNA strand invasion / DNA translocase activity / rDNA binding / nucleosome array spacer activity / RSC-type complex / ATP-dependent chromatin remodeler activity / nucleosome disassembly / SUMOylation of chromatin organization proteins / SWI/SNF complex / nucleosomal DNA binding / nuclear chromosome / NuA4 histone acetyltransferase complex / positive regulation of transcription by RNA polymerase I / histone reader activity / histone H4 reader activity / maturation of LSU-rRNA / : / transcription initiation-coupled chromatin remodeling / cellular response to amino acid starvation / nucleotide-excision repair / chromosome segregation / transcription elongation by RNA polymerase II / helicase activity / double-strand break repair via homologous recombination / chromatin DNA binding / DNA-templated DNA replication / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / structural constituent of chromatin / heterochromatin formation / nucleosome / double-strand break repair / nucleosome assembly / chromatin organization / histone binding / RNA polymerase II-specific DNA-binding transcription factor binding / transcription cis-regulatory region binding / hydrolase activity / chromatin remodeling / protein heterodimerization activity / chromatin binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / positive regulation of DNA-templated transcription / structural molecule activity / positive regulation of transcription by RNA polymerase II / DNA binding / zinc ion binding / nucleoplasm / ATP binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.9 Å | |||||||||
 Authors Authors | Chen ZC / Chen KJ | |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2021 Journal: Cell Discov / Year: 2021Title: Structure of the SWI/SNF complex bound to the nucleosome and insights into the functional modularity. Authors: Zhenyu He / Kangjing Chen / Youpi Ye / Zhucheng Chen /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31137.map.gz emd_31137.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31137-v30.xml emd-31137-v30.xml emd-31137.xml emd-31137.xml | 40.5 KB 40.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31137.png emd_31137.png | 82.7 KB | ||
| Filedesc metadata |  emd-31137.cif.gz emd-31137.cif.gz | 11.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31137 http://ftp.pdbj.org/pub/emdb/structures/EMD-31137 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31137 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31137 | HTTPS FTP |
-Related structure data
| Related structure data |  7egpMC  7eg6C  7egmC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31137.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31137.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The structure of SWI/SNF-nucleosome complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.1484 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : The structure of SWI/SNF chromatin remodeler complex
+Supramolecule #1: The structure of SWI/SNF chromatin remodeler complex
+Supramolecule #2: SWI/SNF
+Supramolecule #3: Histone
+Supramolecule #4: DNA
+Macromolecule #1: SWI/SNF chromatin-remodeling complex subunit SWI1
+Macromolecule #2: SWI/SNF chromatin-remodeling complex subunit SNF5
+Macromolecule #3: SWI/SNF complex subunit SWI3
+Macromolecule #4: Transcription regulatory protein SNF12
+Macromolecule #5: Transcription regulatory protein SNF6
+Macromolecule #6: SWI/SNF global transcription activator complex subunit SWP82
+Macromolecule #7: Transcription regulatory protein SNF2
+Macromolecule #8: Regulator of Ty1 transposition protein 102
+Macromolecule #9: Actin-related protein 7
+Macromolecule #10: Actin-like protein ARP9
+Macromolecule #11: Histone H3.2
+Macromolecule #12: Histone H4
+Macromolecule #13: Histone H2A
+Macromolecule #14: Histone H2B 1.1
+Macromolecule #15: DNA (239-MER)
+Macromolecule #16: DNA (239-MER)
+Macromolecule #17: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #18: BERYLLIUM TRIFLUORIDE ION
+Macromolecule #19: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























