+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30627 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human melatonin receptor MT1 - Gi1 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | gpcr / g-protein / complex / signaling protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationmelatonin receptor activity / Class A/1 (Rhodopsin-like receptors) / mating behavior / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor ...melatonin receptor activity / Class A/1 (Rhodopsin-like receptors) / mating behavior / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / G alpha (q) signalling events / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / hormone binding / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / circadian rhythm / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / ADP signalling through P2Y purinoceptor 12 / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G protein activity / midbody / cell cortex / G alpha (i) signalling events / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Extra-nuclear estrogen signaling / receptor complex / ciliary basal body / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / centrosome / GTP binding / nucleolus / magnesium ion binding / Golgi apparatus / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /    | |||||||||
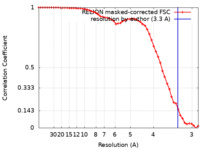
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Okamoto HH / Kusakizako T | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: Cryo-EM structure of the human MT-G signaling complex. Authors: Hiroyuki H Okamoto / Hirotake Miyauchi / Asuka Inoue / Francesco Raimondi / Hirokazu Tsujimoto / Tsukasa Kusakizako / Wataru Shihoya / Keitaro Yamashita / Ryoji Suno / Norimichi Nomura / ...Authors: Hiroyuki H Okamoto / Hirotake Miyauchi / Asuka Inoue / Francesco Raimondi / Hirokazu Tsujimoto / Tsukasa Kusakizako / Wataru Shihoya / Keitaro Yamashita / Ryoji Suno / Norimichi Nomura / Takuya Kobayashi / So Iwata / Tomohiro Nishizawa / Osamu Nureki /    Abstract: Melatonin receptors (MT and MT) transduce inhibitory signaling by melatonin (N-acetyl-5-methoxytryptamine), which is associated with sleep induction and circadian rhythm modulation. Although recently ...Melatonin receptors (MT and MT) transduce inhibitory signaling by melatonin (N-acetyl-5-methoxytryptamine), which is associated with sleep induction and circadian rhythm modulation. Although recently reported crystal structures of ligand-bound MT and MT elucidated the basis of ligand entry and recognition, the ligand-induced MT rearrangement leading to G-coupling remains unclear. Here we report a cryo-EM structure of the human MT-G signaling complex at 3.3 Å resolution, revealing melatonin-induced conformational changes propagated to the G-protein-coupling interface during activation. In contrast to other G-coupled receptors, MT exhibits a large outward movement of TM6, which is considered a specific feature of G-coupled receptors. Structural comparison of G and G complexes demonstrated conformational diversity of the C-terminal entry of the G protein, suggesting loose and variable interactions at the end of the α5 helix of G protein. These notions, together with our biochemical and computational analyses, highlight variable binding modes of Gα and provide the basis for the selectivity of G-protein signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30627.map.gz emd_30627.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30627-v30.xml emd-30627-v30.xml emd-30627.xml emd-30627.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30627_fsc.xml emd_30627_fsc.xml | 5.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_30627.png emd_30627.png | 68.8 KB | ||
| Filedesc metadata |  emd-30627.cif.gz emd-30627.cif.gz | 6.9 KB | ||
| Others |  emd_30627_half_map_1.map.gz emd_30627_half_map_1.map.gz emd_30627_half_map_2.map.gz emd_30627_half_map_2.map.gz | 8.6 MB 8.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30627 http://ftp.pdbj.org/pub/emdb/structures/EMD-30627 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30627 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30627 | HTTPS FTP |
-Related structure data
| Related structure data |  7db6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10714 (Title: human melatonin receptor MT1 - Gi1 complex / Data size: 878.0 EMPIAR-10714 (Title: human melatonin receptor MT1 - Gi1 complex / Data size: 878.0 Data #1: Unaligned movies for MT1-Gi complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30627.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30627.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.44097 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_30627_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
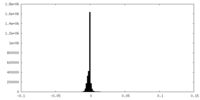
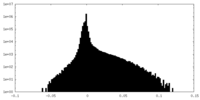
| Density Histograms |
-Half map: #2
| File | emd_30627_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
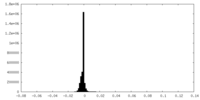
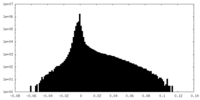
| Density Histograms |
- Sample components
Sample components
-Entire : human melatonin receptor MT1 - Gi1 complex
| Entire | Name: human melatonin receptor MT1 - Gi1 complex |
|---|---|
| Components |
|
-Supramolecule #1: human melatonin receptor MT1 - Gi1 complex
| Supramolecule | Name: human melatonin receptor MT1 - Gi1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.79927 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSQLQSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQD GKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY LSCCRFLDDN Q IVTSSGDT ...String: GSQLQSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQD GKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY LSCCRFLDDN Q IVTSSGDT TCALWDIETG QQTTTFTGHT GDVMSLSLAP DTRLFVSGAC DASAKLWDVR EGMCRQTFTG HESDINAICF FP NGNAFAT GSDDATCRLF DLRADQELMT YSHDNIICGI TSVSFSKSGR LLLAGYDDFN CNVWDALKAD RAGVLAGHDN RVS CLGVTD DGMAVATGSW DSFLKIWN |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.547685 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.720795 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAASSEDL YFQ |
-Macromolecule #5: Melatonin receptor type 1A
| Macromolecule | Name: Melatonin receptor type 1A / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.222805 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKEFMQGN GSALPNASQP VLRGDGARPS WLASALACVL IFTIVVDILG NLLVILSVYR NKKLRNAGN IFVVSLAVAD LVVAIYPYPL VLMSIFNNGW NLGYLHCQVS GFLMGLSVIG SIFNITGIAI NRYCYICHSL K YDKLYSSK ...String: MKTIIALSYI FCLVFADYKD DDDKEFMQGN GSALPNASQP VLRGDGARPS WLASALACVL IFTIVVDILG NLLVILSVYR NKKLRNAGN IFVVSLAVAD LVVAIYPYPL VLMSIFNNGW NLGYLHCQVS GFLMGLSVIG SIFNITGIAI NRYCYICHSL K YDKLYSSK NSLCYVLLIW LLTLAAVLPN LRAGTLQYDP RIYSCTFAQS VSSAYTIAVV VFHFLVPMII VIFCYLRIWI LV LQVRQRV KPDRKPKLKP QDFRNFVTMF VVFVLFAICW APLNFIGLAV ASDPASMVPR IPEWLFVASY YMAYFNSCLN AII YGLLNQ NFRKEYRRII VSLCTARVFF VDSSNDVADR VKWKPSPLMT ENLYFQ UniProtKB: Melatonin receptor type 1A |
-Macromolecule #6: N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}pr...
| Macromolecule | Name: N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide type: ligand / ID: 6 / Number of copies: 1 / Formula: JEV |
|---|---|
| Molecular weight | Theoretical: 259.343 Da |
| Chemical component information | 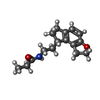 ChemComp-JEV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)