+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30470 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of mammalian NALCN-FAM155A complex at 2.65 angstrom | ||||||||||||||||||
 Map data Map data | map after postprocessing | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | NALCN / Channel / FAM155 / NCA / NLF / TRANSPORT PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationStimuli-sensing channels / positive regulation of synaptic transmission, cholinergic / leak channel activity / regulation of resting membrane potential / voltage-gated sodium channel activity / calcium ion import across plasma membrane / monoatomic ion channel complex / monoatomic cation channel activity / potassium ion transmembrane transport / sodium ion transmembrane transport ...Stimuli-sensing channels / positive regulation of synaptic transmission, cholinergic / leak channel activity / regulation of resting membrane potential / voltage-gated sodium channel activity / calcium ion import across plasma membrane / monoatomic ion channel complex / monoatomic cation channel activity / potassium ion transmembrane transport / sodium ion transmembrane transport / positive regulation of synaptic transmission, GABAergic / calcium ion transmembrane transport / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | ||||||||||||||||||
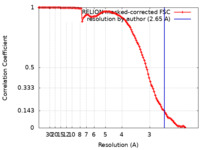
| Method | single particle reconstruction / cryo EM / Resolution: 2.65 Å | ||||||||||||||||||
 Authors Authors | Chen L / Kang Y | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure of voltage-modulated sodium-selective NALCN-FAM155A channel complex. Authors: Yunlu Kang / Jing-Xiang Wu / Lei Chen /  Abstract: Resting membrane potential determines the excitability of the cell and is essential for the cellular electrical activities. The NALCN channel mediates sodium leak currents, which positively adjust ...Resting membrane potential determines the excitability of the cell and is essential for the cellular electrical activities. The NALCN channel mediates sodium leak currents, which positively adjust resting membrane potential towards depolarization. The NALCN channel is involved in several neurological processes and has been implicated in a spectrum of neurodevelopmental diseases. Here, we report the cryo-EM structure of rat NALCN and mouse FAM155A complex to 2.7 Å resolution. The structure reveals detailed interactions between NALCN and the extracellular cysteine-rich domain of FAM155A. We find that the non-canonical architecture of NALCN selectivity filter dictates its sodium selectivity and calcium block, and that the asymmetric arrangement of two functional voltage sensors confers the modulation by membrane potential. Moreover, mutations associated with human diseases map to the domain-domain interfaces or the pore domain of NALCN, intuitively suggesting their pathological mechanisms. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30470.map.gz emd_30470.map.gz | 49.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30470-v30.xml emd-30470-v30.xml emd-30470.xml emd-30470.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30470_fsc.xml emd_30470_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_30470.png emd_30470.png | 79.8 KB | ||
| Filedesc metadata |  emd-30470.cif.gz emd-30470.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30470 http://ftp.pdbj.org/pub/emdb/structures/EMD-30470 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30470 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30470 | HTTPS FTP |
-Related structure data
| Related structure data |  7cu3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30470.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30470.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map after postprocessing | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : NALCN-FAM155A complex
| Entire | Name: NALCN-FAM155A complex |
|---|---|
| Components |
|
-Supramolecule #1: NALCN-FAM155A complex
| Supramolecule | Name: NALCN-FAM155A complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium leak channel non-selective protein
| Macromolecule | Name: Sodium leak channel non-selective protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200.682031 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLKRKQSSRV EAQPVTDFGP DESLSDNADI LWINKPWVHS LLRICAIISV ISVCMNTPMT FEHYPPLQYV TFTLDTLLMF LYTAEMIAK MHIRGIVKGD SSYVKDRWCV FDGFMVFCLW VSLVLQVFEI ADIVDQMSPW GMLRIPRPLI MIRAFRIYFR F ELPRTRIT ...String: MLKRKQSSRV EAQPVTDFGP DESLSDNADI LWINKPWVHS LLRICAIISV ISVCMNTPMT FEHYPPLQYV TFTLDTLLMF LYTAEMIAK MHIRGIVKGD SSYVKDRWCV FDGFMVFCLW VSLVLQVFEI ADIVDQMSPW GMLRIPRPLI MIRAFRIYFR F ELPRTRIT NILKRSGEQI WSVSIFLLFF LLLYGILGVQ MFGTFTYHCV VNDTKPGNVT WNSLAIPDTH CSPELEEGYQ CP PGFKCMD LEDLGLSRQE LGYSGFNEIG TSIFTVYEAS SQEGWVFLMY RAIDSFPRWR SYFYFITLIF FLAWLVKNVF IAV IIETFA EIRVQFQQMW GTRSSTTSTA TTQMFHEDAA GGWQLVAVDV NKPQGRAPAC LQKMMRSSVF HMFILSMVTV DVIV AASNY YKGENFRRQY DEFYLAEVAF TVLFDLEALL KIWCLGFTGY ISSSLHKFEL LLVIGTTLHV YPDLYHSQFT YFQVL RVVR LIKISPALED FVYKIFGPGK KLGSLVVFTA SLLIVMSAIS LQMFCFVEEL DRFTTFPRAF MSMFQILTQE GWVDVM DQT LNAVGHMWAP LVAIYFILYH LFATLILLSL FVAVILDNLE LDEDLKKLKQ LKQSEANADT KEKLPLRLRI FEKFPNR PQ MVKISKLPSD FTVPKIRESF MKQFIDRQQQ DTCCLFRILP STSSSSCDNP KRPTVEDNKY IDQKLRKSVF SIRARNLL E KETAVTKILR ACTRQRMLSG SFEGQPAKER SILSVQHHIR QERRSLRHGS NSQRISRGKS LETLTQDHSN TVRYRNAQR EDSEIKMIQE KKEQAEMKRK VQEEELRENH PYFDKPLFIV GREHRFRNFC RVVVRARFNA SKTDPVTGAV KNTKYHQLYD LLGLVTYLD WVMITVTICS CISMMFESPF RRVMHAPTLQ IAEYVFVIFM SIELNLKIMA DGLFFTPTAV IRDFGGVMDI F IYLVSLIF LCWMPQNVPA ESGAQLLMVL RCLRPLRIFK LVPQMRKVVR ELFSGFKEIF LVSILLLTLM LVFASFGVQL FA GKLAKCN DPNIIRREDC NGIFRINVSV SKNLNLKLRP GEKKPGFWVP RVWANPRNFN FDNVGNAMLA LFEVLSLKGW VEV RDVIIH RVGPIHGIYI HVFVFLGCMI GLTLFVGVVI ANFNENKGTA LLTVDQRRWE DLKSRLKIAQ PLHLPPRPDN DGFR AKMYD ITQHPFFKRT IALLVLAQSV LLSVKWDVED PVTVPLATMS VVFTFIFVLE VTMKIIAMSP AGFWQSRRNR YDLLV TSLG VVWVVLHFAL LNAYTYMMGA CVIVFRFFSI CGKHVTLKML LLTVVVSMYK SFFIIVGMFL LLLCYAFAGV VLFGTV KYG ENINRHANFS SAGKAITVLF RIVTGEDWNK IMHDCMVQPP FCTPDEFTYW ATDCGNYAGA LMYFCSFYVI IAYIMLN LL VAIIVENFSL FYSTEEDQLL SYNDLRHFQI IWNMVDDKRE GVIPTFRVKF LLRLLRGRLE VDLDKDKLLF KHMCYEME R LHNGGDVTFH DVLSMLSYRS VDIRKSLQLE ELLAREQLEY TIEEEVAKQT IRMWLKKCLK RIRAKQQQSC SIIHSLRES QQQELSRFLN PPSIETTQPS EDTNANSQDH NTQPESSSQQ QLLSPTLSDR GGSRQDAADT GKPQRKIGQW RLPSAPKPIS HSVSSVNLR FGGRTTMKSV VCKMNPMPDT ASCGSEVKKW WTRQLTVESD ESGDDLLDI UniProtKB: Sodium leak channel NALCN |
-Macromolecule #2: Transmembrane protein FAM155A
| Macromolecule | Name: Transmembrane protein FAM155A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.795801 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTRGAWMCRQ YDDGLKIWLA APRENEKPFI DSERAQKWRL SLASLLFFTV LLSDHLWFCA EAKLTRTRDK EHHQQQQQQQ QQQQQQQQQ QQQQQQRQQQ RQRQQQRQRQ QEPSWPALLA SMGESSPAAQ AHRLLSASSS PTLPPSPGGG GGSKGNRGKN N RSRALFLG ...String: MTRGAWMCRQ YDDGLKIWLA APRENEKPFI DSERAQKWRL SLASLLFFTV LLSDHLWFCA EAKLTRTRDK EHHQQQQQQQ QQQQQQQQQ QQQQQQRQQQ RQRQQQRQRQ QEPSWPALLA SMGESSPAAQ AHRLLSASSS PTLPPSPGGG GGSKGNRGKN N RSRALFLG NSAKPVWRLE TCYPQGASSG QCFTVESADA VCARNWSRGA AAGEEQSSRG SRPTPLWNLS DFYLSFCNSY TL WELFSGL SSPSTLNCSL DVVLTEGGEM TTCRQCIEAY QDYDHHAQEK YEEFESVLHK YLQSDEYSVK SCPEDCKIVY KAW LCSQYF EVTQFNCRKT IPCKQYCLEV QTRCPFILPD NDEVIYGGLS SFICTGLYET FLTNDEPECC DIRSEEQTAP RPKG TVDRR DSCPRTSLTV SSATRLCPGR LKLCVLVLIL LHTVLTASAA QNSTGLGLGG LPTLEDNSTR ED UniProtKB: NALCN channel auxiliary factor 1 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyl...
| Macromolecule | Name: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate type: ligand / ID: 4 / Number of copies: 12 / Formula: 6OU |
|---|---|
| Molecular weight | Theoretical: 717.996 Da |
| Chemical component information |  ChemComp-6OU: |
-Macromolecule #5: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 5 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)