+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Initial DNA-lesion (Cy5) binding by XPC and TFIIH focused | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | ||||||||||||
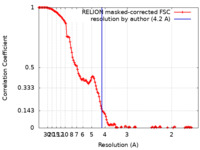
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | ||||||||||||
 Authors Authors | Kim J / Yang W | ||||||||||||
| Funding support |  United States, United States,  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Lesion recognition by XPC, TFIIH and XPA in DNA excision repair. Authors: Jinseok Kim / Chia-Lung Li / Xuemin Chen / Yanxiang Cui / Filip M Golebiowski / Huaibin Wang / Fumio Hanaoka / Kaoru Sugasawa / Wei Yang /     Abstract: Nucleotide excision repair removes DNA lesions caused by ultraviolet light, cisplatin-like compounds and bulky adducts. After initial recognition by XPC in global genome repair or a stalled RNA ...Nucleotide excision repair removes DNA lesions caused by ultraviolet light, cisplatin-like compounds and bulky adducts. After initial recognition by XPC in global genome repair or a stalled RNA polymerase in transcription-coupled repair, damaged DNA is transferred to the seven-subunit TFIIH core complex (Core7) for verification and dual incisions by the XPF and XPG nucleases. Structures capturing lesion recognition by the yeast XPC homologue Rad4 and TFIIH in transcription initiation or DNA repair have been separately reported. How two different lesion recognition pathways converge and how the XPB and XPD helicases of Core7 move the DNA lesion for verification are unclear. Here we report on structures revealing DNA lesion recognition by human XPC and DNA lesion hand-off from XPC to Core7 and XPA. XPA, which binds between XPB and XPD, kinks the DNA duplex and shifts XPC and the DNA lesion by nearly a helical turn relative to Core7. The DNA lesion is thus positioned outside of Core7, as would occur with RNA polymerase. XPB and XPD, which track the lesion-containing strand but translocate DNA in opposite directions, push and pull the lesion-containing strand into XPD for verification. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29674.map.gz emd_29674.map.gz | 11.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29674-v30.xml emd-29674-v30.xml emd-29674.xml emd-29674.xml | 33.5 KB 33.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29674_fsc.xml emd_29674_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_29674.png emd_29674.png | 70.6 KB | ||
| Masks |  emd_29674_msk_1.map emd_29674_msk_1.map | 216 MB |  Mask map Mask map | |
| Others |  emd_29674_half_map_1.map.gz emd_29674_half_map_1.map.gz emd_29674_half_map_2.map.gz emd_29674_half_map_2.map.gz | 153.2 MB 153.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29674 http://ftp.pdbj.org/pub/emdb/structures/EMD-29674 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29674 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29674 | HTTPS FTP |
-Validation report
| Summary document |  emd_29674_validation.pdf.gz emd_29674_validation.pdf.gz | 730.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29674_full_validation.pdf.gz emd_29674_full_validation.pdf.gz | 729.8 KB | Display | |
| Data in XML |  emd_29674_validation.xml.gz emd_29674_validation.xml.gz | 21 KB | Display | |
| Data in CIF |  emd_29674_validation.cif.gz emd_29674_validation.cif.gz | 27.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29674 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29674 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29674 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29674 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29674.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29674.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29674_msk_1.map emd_29674_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29674_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29674_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : bulky DNA lesion recognition complex 1
+Supramolecule #1: bulky DNA lesion recognition complex 1
+Macromolecule #1: General transcription and DNA repair factor IIH helicase subunit XPB
+Macromolecule #2: TFIIH basal transcription factor complex helicase XPD subunit
+Macromolecule #3: General transcription factor IIH subunit 1
+Macromolecule #4: General transcription factor IIH subunit 4
+Macromolecule #5: General transcription factor IIH subunit 2
+Macromolecule #6: General transcription factor IIH subunit 3
+Macromolecule #7: General transcription factor IIH subunit 5
+Macromolecule #8: DNA repair protein complementing XP-C cells
+Macromolecule #9: UV excision repair protein RAD23 homolog B
+Macromolecule #10: Centrin-2
+Macromolecule #11: DNA1
+Macromolecule #12: DNA2
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6243 / Average exposure time: 2.5 sec. / Average electron dose: 54.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: correlation coefficient |
|---|
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)